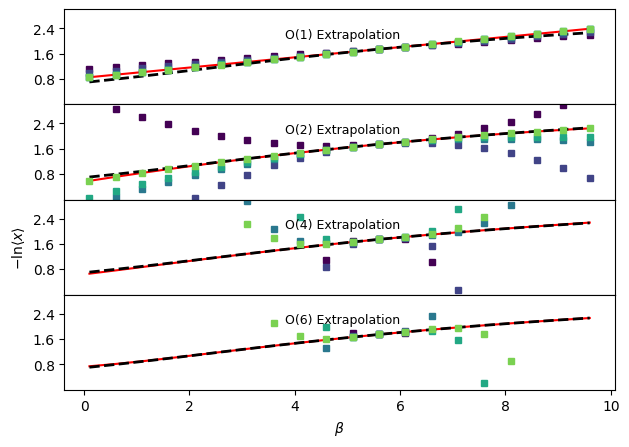
Temperature independent negative logarithm of average observable#
This type of extrapolation is useful for extrapolating hard-sphere chemical potentials in temperature or volume, or other quantities involving logarithms of probabilities (e.g. PMFs).
This is in fact easier than Case 2 because we do not need to augment the data in any way - we just set a flag that the quantity to extrapolate is the negative logarithm of an ensemble average, i.e., post_func = 'minus_log'. Note, though, that we handled the -log calculation in the definition of the derivatives (even at zeroth order). This means we want to just pass data, not the -log of the data. Also note that post_func can take any function utilizing sympy expressions for modifying the average observable we want to extrapolate, so passing post_func = lambda x: -sympy.log(x) would be equivalent. If you’re applying the function to the observable itself, that’s just a different observable and you don’t need to worry about what we’re doing in this notebook… it’s the difference between \(\ln \langle x \rangle\) and \(\langle \ln x \rangle\).
%matplotlib inline
import cmomy
import matplotlib as mpl
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
# Import idealgas module
from thermoextrap import idealgas
rng = cmomy.random.default_rng(seed=0)
# Define test betas and reference beta
betas = np.arange(0.1, 10.0, 0.5)
beta_ref = betas[11]
vol = 1.0
# Define orders to extrapolate to
orders = [1, 2, 4, 6]
order = orders[-1]
npart = 1000 # Number of particles (in single configuration)
nconfig = 100_000 # Number of configurations
# Generate all the data we could want
xdata, udata = idealgas.generate_data((nconfig, npart), beta_ref, vol)
import thermoextrap as xtrap
# Create and train extrapolation model
xem_log = xtrap.beta.factory_extrapmodel(
beta=beta_ref,
post_func="minus_log",
data=xtrap.DataCentralMomentsVals.from_vals(
order=orders[-1],
xv=xr.DataArray(xdata, dims="rec"),
uv=xr.DataArray(udata, dims="rec"),
deriv_dim=None,
central=True,
),
)
# Check the derivatives
print("Model parameters (derivatives):")
print(xem_log.derivs(norm=False))
print("\n")
# Finally, look at predictions
print("Model predictions:")
print(xem_log.predict(betas[:4], order=2))
print("\n")
Model parameters (derivatives):
<xarray.DataArray (order: 7)> Size: 56B
array([ 1.7438, 0.1615, -0.0186, 0.015 , 0.612 , -1.9426, -1.9077])
Dimensions without coordinates: order
Model predictions:
<xarray.DataArray (beta: 4)> Size: 32B
array([0.5741, 0.7037, 0.8286, 0.9489])
Coordinates:
* beta (beta) float64 32B 0.1 0.6 1.1 1.6
dalpha (beta) float64 32B -5.5 -5.0 -4.5 -4.0
beta0 float64 8B 5.6
# And bootstrapped uncertainties
print("Bootstrapped uncertainties in predictions:")
print(xem_log.resample(sampler={"nrep": 100}).predict(betas[:4], order=3).std("rep"))
Bootstrapped uncertainties in predictions:
<xarray.DataArray (beta: 4)> Size: 32B
array([2.0496, 1.54 , 1.1227, 0.7887])
Coordinates:
* beta (beta) float64 32B 0.1 0.6 1.1 1.6
dalpha (beta) float64 32B -5.5 -5.0 -4.5 -4.0
beta0 float64 8B 5.6
# Repeat comparison of results, but for -ln<x> instead of <x>
fig, ax = plt.subplots(len(orders), sharex=True, sharey=True)
nsampvals = np.array((10.0 * np.ones(5)) ** np.arange(1, 6), dtype=int)
nsampcolors = plt.cm.viridis(np.arange(0.0, 1.0, float(1.0 / len(nsampvals))))
# First plot the analytical result
for a in ax:
a.plot(betas, -np.log(idealgas.x_ave(betas, vol)), "k--", linewidth=2.0)
# Next look at extrapolation with an infinite number of samples
# This is possible in the ideal gas model in both temperature and volume
for j, o in enumerate(orders):
trueExtrap, trueDerivs = idealgas.x_beta_extrap_minuslog(o, beta_ref, betas, vol)
ax[j].plot(betas, trueExtrap, "r-", zorder=0)
if j == len(orders) - 1:
print(f"True extrapolation coefficients: {trueDerivs}")
for i, n in enumerate(nsampvals):
thisinds = rng.choice(len(xdata), size=n, replace=False)
# Get parameters for extrapolation model with this data by training it - the parameters are the derivatives
xem_log = xtrap.beta.factory_extrapmodel(
beta=beta_ref,
post_func="minus_log",
data=xtrap.DataCentralMomentsVals.from_vals(
order=orders[-1],
uv=xr.DataArray(udata[thisinds], dims="rec"),
xv=xr.DataArray(xdata[thisinds], dims="rec"),
central=True,
),
)
out = xem_log.predict(betas, cumsum=True)
print(
f"\t With N_configs = {n}: {xem_log.derivs(norm=False).values.flatten()}"
) # Have to flatten because observable is 1-D
for j, o in enumerate(orders):
out.sel(order=o).plot(
marker="s",
ms=4,
color=nsampcolors[i],
ls="None",
label=f"N={n}",
ax=ax[j],
)
ax[2].set_ylabel(r"$-\mathrm{ln} \langle x \rangle$")
ax[-1].set_xlabel(r"$\beta$")
for j, o in enumerate(orders):
ax[j].annotate(
f"O({o}) Extrapolation", xy=(0.4, 0.7), xycoords="axes fraction", fontsize=9
)
ax[0].set_ylim((0.0, 3.0))
ax[-1].yaxis.set_major_locator(mpl.ticker.MaxNLocator(nbins=4, prune="both"))
fig.tight_layout()
fig.subplots_adjust(hspace=0.0)
for a in ax:
a.set_title(None)
plt.show()
True extrapolation coefficients: [ 1.7438e+00 1.6106e-01 -1.7727e-02 3.6676e-04 2.1114e-03 -1.5377e-03
3.9927e-04]
With N_configs = 10: [ 1.7469e+00 1.1364e-01 1.3435e-01 -8.7672e-01 -1.8012e+01 -1.0596e+02
3.2007e+03]
With N_configs = 100: [ 1.7423e+00 1.3459e-01 -1.9823e-01 1.2182e+00 -1.0755e+01 -6.0855e+01
3.2539e+03]
With N_configs = 1000: [ 1.7426 0.1588 -0.0696 -0.1739 0.8373 46.7624 56.4464]
With N_configs = 10000: [ 1.7436 0.161 -0.0531 -0.2102 4.1724 -23.4454 22.6547]
With N_configs = 100000: [ 1.7438 0.1615 -0.0186 0.015 0.612 -1.9426 -1.9077]