Quickstart#
Atoms Data#
Atoms is the primary data structure for atomistic modeling in NFFLr.
This represents an atomistic system in the same way as ase:
a \(3 \times 3\)
cellmatrixan \(N \times 3\) coordinates array
positionsan array of
Natomicnumbers
These variables are stored as PyTorch tensors to facilitate auto-batching, flexible conversion to graph deep learning formats, and automatic differentiation.
import torch
import nfflr
cell = 4.1 * torch.eye(3)
scaled_positions = torch.tensor([[0,0,0], [0.5, 0.5, 0.5]])
numbers = torch.tensor([24, 22])
atoms = nfflr.Atoms(cell, scaled_positions @ cell, numbers)
atoms
/opt/hostedtoolcache/Python/3.10.13/x64/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
2024-03-08 20:08:26,588 INFO util.py:154 -- Missing packages: ['ipywidgets']. Run `pip install -U ipywidgets`, then restart the notebook server for rich notebook output.
2024-03-08 20:08:26,739 INFO util.py:154 -- Missing packages: ['ipywidgets']. Run `pip install -U ipywidgets`, then restart the notebook server for rich notebook output.
Atoms(cell=tensor([[4.1000, 0.0000, 0.0000],
[0.0000, 4.1000, 0.0000],
[0.0000, 0.0000, 4.1000]]), positions=tensor([[0.0000, 0.0000, 0.0000],
[2.0500, 2.0500, 2.0500]]), numbers=tensor([24, 22]), _batch_num_atoms=None)
from ase import Atoms as AseAtoms
from ase.visualize.plot import plot_atoms
import matplotlib.pyplot as plt
%matplotlib inline
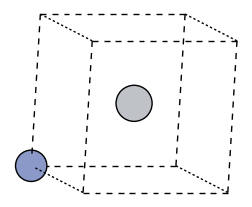
fig, ax = plt.subplots(figsize=(3,3))
ase_atoms = nfflr.to_ase(atoms)
plot_atoms(ase_atoms, ax, radii=0.3, rotation=("10x,20y,0z"), show_unit_cell=2)
ax.axis("off");
Models#
Common model interface#
NFFLr models are PyTorch modules for modeling properties of atomistic systems.
Different modeling approaches use a variety of input representations;
NFFLr aims to simplify exploratory research by providing a common modeling interface, where all models internally transform Atoms inputs to their native input representation, e.g. the DGLGraph structure expected by ALIGNN.
cfg = nfflr.models.ALIGNNConfig(alignn_layers=2, gcn_layers=2)
alignn_model = nfflr.models.ALIGNN(cfg)
with torch.no_grad():
print(alignn_model(atoms))
warning: could not load CGCNN features for 103
Setting it to max atomic number available here, 103
warning: could not load CGCNN features for 101
Setting it to max atomic number available here, 103
warning: could not load CGCNN features for 102
Setting it to max atomic number available here, 103
tensor(-0.3866)
NFFLr also transparently converts inputs from common atomistic modeling libraries, such as jarvis and ase.
This is an experimental feature that is currently implemented with the plum multiple dispatch library.
For example, calling alignn_model on an ase.Atoms structure automatically converts the data to nfflr.Atoms and then to the ALIGNN DGLGraph format:
with torch.no_grad():
print(alignn_model(ase_atoms))
convert
tensor(-0.3866)
Force field models#
Enabling the compute_forces model configuration field will cause the model to compute both a scalar property prediction
and its (negative) gradient with respect to the (cartesian) atomic coordinates - i.e. the force components on each atom.
cfg = nfflr.models.ALIGNNConfig(
alignn_layers=2,
gcn_layers=2,
compute_forces=True
)
alignn_model = nfflr.models.ALIGNN(cfg)
alignn_model(atoms)
{'energy': tensor(-0.3729, grad_fn=<SqueezeBackward0>),
'forces': tensor([[ 2.6822e-07, -1.6578e-07, 6.9477e-07],
[-2.8312e-07, 2.1793e-07, -6.4261e-07]], grad_fn=<MulBackward0>),
'stress': tensor([[[ 1.3659e+00, 1.2666e-07, 2.8312e-07],
[ 2.0117e-07, 1.3659e+00, -1.6382e-06],
[ 3.6508e-07, -1.6605e-06, 1.3659e+00]]],
grad_fn=<SegmentReduceBackward>)}
input representations#
NFFLr also allows to directly provide inputs to models in the native input representation expected by the model.
This facilitates efficient precomputation and caching during training.
For example, ALIGNN requires DGLGraph inputs with node features atomic_number and edge features r (the bond vectors pointing from atoms to their neighbors).
from nfflr.data.graph import periodic_radius_graph
g = periodic_radius_graph(atoms, r=6)
g
Graph(num_nodes=2, num_edges=52,
ndata_schemes={'coord': Scheme(shape=(3,), dtype=torch.float32), 'atomic_number': Scheme(shape=(), dtype=torch.int32)}
edata_schemes={'r': Scheme(shape=(3,), dtype=torch.float32)})
alignn_model(g)
{'energy': tensor(-0.3757, grad_fn=<SqueezeBackward0>),
'forces': tensor([[ 2.6226e-06, -7.1526e-07, 2.0565e-06],
[-2.6226e-06, 5.9605e-07, -2.0265e-06]], grad_fn=<MulBackward0>),
'stress': tensor([[[-6.0826e+00, 8.3447e-07, -7.1526e-07],
[ 8.3447e-07, -6.0826e+00, -4.7684e-07],
[-7.1526e-07, -4.7684e-07, -6.0826e+00]]],
grad_fn=<SegmentReduceBackward>)}
Training utilities#
AtomsDataset#
NFFLr makes it easy to load data and transform it into various formats.
The primary ways of interacting with data are Atoms and AtomsDataset,
which is a PyTorch DataSet that returns Atoms instances.
The most convenient way to get started is with a named Jarvis dataset:
dataset = nfflr.AtomsDataset("dft_3d", target="formation_energy_peratom")
dataset_name='dft_3d'
Obtaining 3D dataset 76k ...
Reference:https://www.nature.com/articles/s41524-020-00440-1
Other versions:https://doi.org/10.6084/m9.figshare.6815699
Loading the zipfile...
Loading completed.
The dataset yields a tuple of an Atoms instance and the target value, e.g., target="formation_energy_peratom":
atoms, target = dataset[0]
print(f"{atoms.cell=}")
print(f"{atoms.positions=}")
print(f"{atoms.numbers=}")
print(f"{target=}")
atoms.cell=tensor([[3.5669, 0.0000, -0.0000],
[0.0000, 3.5669, -0.0000],
[-0.0000, -0.0000, 9.3971]])
atoms.positions=tensor([[2.6752, 2.6752, 7.3761],
[0.8917, 0.8917, 2.0210],
[0.8917, 2.6752, 4.6985],
[2.6752, 0.8917, 4.6985],
[0.8917, 2.6752, 0.0000],
[2.6752, 0.8917, 0.0000],
[2.6752, 2.6752, 2.8895],
[0.8917, 0.8917, 6.5076]])
atoms.numbers=tensor([22, 22, 29, 29, 14, 14, 33, 33], dtype=torch.int32)
target=tensor(-0.4276)
Internally, AtomsDataset uses a pandas dataframe to store the datasets, so any key in the jarvis dataset is a valid target.
For example, dft_3d contains a large number of target properties, including some non-scalar quantities:
selected_cols = ("jid", "formula", "formation_energy_peratom", "optb88vdw_bandgap", "elastic_tensor")
dataset.df.loc[:,selected_cols].head()
| jid | formula | formation_energy_peratom | optb88vdw_bandgap | elastic_tensor | |
|---|---|---|---|---|---|
| 0 | JVASP-90856 | TiCuSiAs | -0.42762 | 0.000 | na |
| 1 | JVASP-86097 | DyB6 | -0.41596 | 0.000 | na |
| 2 | JVASP-64906 | Be2OsRu | 0.04847 | 0.000 | na |
| 3 | JVASP-98225 | KBi | -0.44140 | 0.472 | na |
| 4 | JVASP-10 | VSe2 | -0.71026 | 0.000 | [[136.4, 27.8, 17.5, 0.0, -5.5, 0.0], [27.8, 1... |
We can change the target column, but missing values currently need to be handled manually.
dataset.target = "elastic_tensor"
atoms, elastic_tensor = dataset[4]
elastic_tensor
tensor([[136.4000, 27.8000, 17.5000, 0.0000, -5.5000, 0.0000],
[ 27.8000, 136.4000, 17.5000, 0.0000, 5.5000, 0.0000],
[ 17.5000, 17.5000, 40.7000, 0.0000, 0.0000, 0.0000],
[ 0.0000, 0.0000, 0.0000, 54.3000, 0.0000, -5.5000],
[ -5.5000, 5.5000, 0.0000, 0.0000, 13.7000, 0.0000],
[ 0.0000, 0.0000, 0.0000, -5.5000, 0.0000, 13.7000]])
Force field datasets#
Force field datasets like mlearn, alignn_ff_db, and m3gnet have a special target key target="energy_and_forces" that configure AtomsDataset to return a dictionary of target values containing the total energy of the atomic configuration, the forces, and the stresses if they are available.
dataset = nfflr.AtomsDataset("mlearn", target="energy_and_forces")
atoms, target = dataset[0]
target
dataset_name='mlearn'
Obtaining mlearn dataset 1730...
Reference:https://github.com/materialsvirtuallab/mlearn
0%| | 0.00/2.57M [00:00<?, ?iB/s]
2%|▏ | 52.2k/2.57M [00:00<00:06, 379kiB/s]
9%|▉ | 226k/2.57M [00:00<00:02, 889kiB/s]
39%|███▊ | 992k/2.57M [00:00<00:00, 2.97MiB/s]
100%|██████████| 2.57M/2.57M [00:00<00:00, 5.15MiB/s]
Loading the zipfile...
Loading completed.
{'energy': tensor(-604.2623),
'forces': tensor([[-1.9282e-01, -1.8793e+00, -6.6374e-01],
[-8.2543e-03, -2.0313e-01, 3.6808e-01],
[-5.5372e-01, -1.4736e+00, 1.2997e+00],
[ 4.5678e-01, 5.1175e-01, -1.0934e+00],
[-1.6499e+00, -1.6259e+00, 4.5255e-01],
[-1.6698e-01, 6.8080e-01, 6.7749e-01],
[ 3.6802e-02, -3.1423e+00, -2.0166e+00],
[-1.0730e-01, -3.5780e-01, 1.1357e+00],
[-1.9132e-01, 5.1381e-01, 3.4296e-01],
[ 2.0090e+00, 1.5143e+00, -3.5578e-01],
[-1.7128e-01, -2.7808e+00, -1.4215e+00],
[-9.3987e-01, -1.6757e-02, 7.9322e-01],
[ 3.7190e-01, -9.0627e-01, -5.2933e-01],
[ 5.6458e-01, -9.6833e-01, -7.0043e-01],
[-4.5756e-01, -6.5868e-02, -3.7038e-01],
[-1.2044e+00, 6.3979e-01, 7.5036e-01],
[-1.5743e+00, 6.4479e-02, -6.7272e-01],
[-9.8223e-01, -9.5903e-02, -8.7198e-01],
[ 4.9518e-01, -2.7982e-01, -4.6208e-01],
[ 3.3000e-01, 1.7643e-01, 2.0947e+00],
[ 3.3517e-01, 1.4522e+00, 3.6359e-01],
[-4.4930e-01, -3.1648e-01, 2.1246e-01],
[-5.8361e-01, 1.0337e+00, -1.0099e+00],
[ 1.4334e+00, 1.4563e+00, 4.8775e-01],
[-1.2193e+00, -1.8368e-01, 1.7678e-01],
[-1.8822e-02, -3.3724e-01, 5.0373e-01],
[ 9.7925e-01, 3.4629e-01, 2.7126e-01],
[ 1.3972e+00, 1.0313e-01, 2.1936e+00],
[ 1.4154e+00, 1.0657e+00, 5.6893e-01],
[-5.3909e-01, 6.2667e-01, 7.9585e-01],
[-8.0468e-02, 9.3723e-01, -1.7657e+00],
[ 6.4826e-01, 1.3950e-03, -1.1809e+00],
[ 1.7236e+00, 5.0571e-01, 2.0909e-01],
[-6.3469e-01, 3.2798e+00, 1.3690e+00],
[-2.8363e-01, 1.3372e+00, -3.8005e-01],
[-1.0848e+00, -5.7622e-01, -6.1141e-01],
[-1.8884e+00, 5.1697e-01, -1.0889e-01],
[-5.3894e-01, 2.1740e+00, 2.2013e+00],
[ 1.5727e+00, -9.5217e-01, 9.6934e-01],
[ 3.8191e-01, 3.4829e-01, 1.2664e+00],
[-1.1411e+00, 1.2328e+00, 1.2866e+00],
[ 1.1776e+00, 7.2366e-01, -1.5056e+00],
[-1.3455e+00, -4.8714e-01, 4.1776e-01],
[ 2.7808e-01, -1.4488e-01, 1.2792e+00],
[-2.0664e-01, 1.4243e+00, 1.2686e+00],
[ 1.3897e+00, 7.7333e-01, -8.4011e-01],
[-7.0459e-01, -2.1634e+00, 1.0630e+00],
[-9.9009e-01, -6.2214e-01, -9.4072e-03],
[ 3.3802e-01, 3.1611e-01, 1.3336e-01],
[-1.2308e+00, -2.7998e-01, -9.0719e-01],
[ 1.5169e+00, -6.4886e-01, -1.4431e+00],
[ 2.3966e+00, 1.3065e+00, 3.9503e-01],
[ 4.8711e-01, 2.6996e-03, 5.6954e-01],
[ 3.0038e-02, 9.8048e-01, 9.6736e-02],
[-2.8896e-01, 6.9839e-01, 1.1865e-01],
[-7.0303e-01, 1.5889e+00, 1.0517e+00],
[ 1.4835e+00, -7.5193e-01, -4.8107e-01],
[ 4.3507e-01, -7.6680e-01, -7.6512e-01],
[ 1.6324e+00, -9.0497e-01, -1.7391e-01],
[-7.7163e-01, 8.8480e-01, -1.0546e-01],
[ 1.5508e+00, -1.4519e-01, -6.3183e-01],
[ 1.4062e+00, 4.8017e-01, 2.4209e-01],
[-8.2076e-01, -1.1055e+00, -3.7652e-01],
[-1.7866e+00, -1.0725e-01, -7.5774e-01],
[ 6.6219e-01, -1.1061e+00, 6.6820e-01],
[ 4.5689e-01, -3.1297e-01, 5.2079e-01],
[-2.3750e-01, 1.6904e+00, -7.2430e-01],
[ 1.5449e+00, 1.4885e+00, -5.6164e-01],
[ 1.6403e+00, -1.3929e+00, -1.3473e-01],
[-5.0026e-01, -7.1965e-01, -6.3690e-01],
[ 1.8875e-01, -8.0416e-01, 1.0578e+00],
[ 7.4767e-01, -2.7263e-01, 1.0396e-01],
[ 1.0797e+00, 6.2834e-01, -1.0441e+00],
[-9.1592e-01, -1.0053e+00, -1.6651e-01],
[-2.4538e-01, 1.1315e+00, -2.5051e-01],
[-2.6349e-01, -3.9915e-01, 5.2209e-01],
[ 8.3324e-01, 2.9588e-02, 4.1156e-01],
[ 1.3736e-01, 5.2689e-01, -7.6983e-01],
[ 1.8699e+00, -5.6415e-01, -1.2089e+00],
[-8.2056e-01, -5.2394e-01, -1.0657e-01],
[-1.3969e-01, -2.1350e-01, 2.1012e-01],
[-8.5827e-01, -2.9145e-01, -8.8987e-02],
[-2.7861e-01, -6.4112e-01, 2.7514e-01],
[-7.0377e-01, -1.6119e-01, -1.6974e-02],
[-4.9227e-01, -5.5502e-01, -1.6419e+00],
[ 1.3265e+00, 5.1135e-01, -2.0431e-01],
[-6.3025e-01, -4.0777e-01, -7.4116e-01],
[-2.7982e+00, -8.6561e-01, 7.2870e-01],
[ 4.4176e-01, -6.1487e-01, -1.5266e+00],
[-8.2469e-01, -1.5254e+00, 2.2129e-01],
[-4.1837e-01, 4.5957e-01, -9.3009e-01],
[-1.3448e+00, -3.8741e-01, 5.7946e-01],
[-3.5803e-02, -4.9431e-01, -3.3611e-01],
[ 1.3890e+00, -2.3396e-01, -5.8913e-01],
[ 4.6561e-01, -1.6739e+00, -5.8580e-01],
[-5.4732e-02, 1.2076e+00, -6.2845e-01],
[-1.9202e+00, 2.6483e-01, -4.7163e-01],
[ 2.3382e-01, -1.9371e-01, 8.8642e-01],
[-5.4136e-02, 7.5257e-01, -7.5428e-01],
[-1.2954e+00, -8.2409e-01, -2.3798e-01],
[ 2.2413e-01, -5.5878e-02, -5.6709e-01],
[ 1.0508e+00, 4.7083e-01, 1.0494e+00],
[ 1.1418e+00, 3.9075e-01, 2.2798e-01],
[-1.6860e+00, 8.3186e-01, 7.9992e-01],
[-1.1271e+00, 7.7508e-02, 9.2828e-01],
[-1.0157e+00, 5.2795e-01, -1.9179e-01],
[ 4.6428e-01, -1.5829e-01, 7.1079e-01]]),
'volume': tensor(1165.6177)}