ADEPT 2-State¶
This algorithm limits the generalized algorithm for state-detection [] to cases with a single state as seen in the figure below. This simplified approach speeds up the analysis considerably and is appropriate to use for many applications, for example the detection of PEG, small molecules, DNA homopolymers, etc. The adept2State class uses a simplified form of the expression for the ionic current across a nanopore as shown below. Settings that control the fit are defined through the settings file and are described in more detail in the Optimizing Settings section. This functional form is fit to a time-series from a single event to recover optimal parameters for the mdoel.
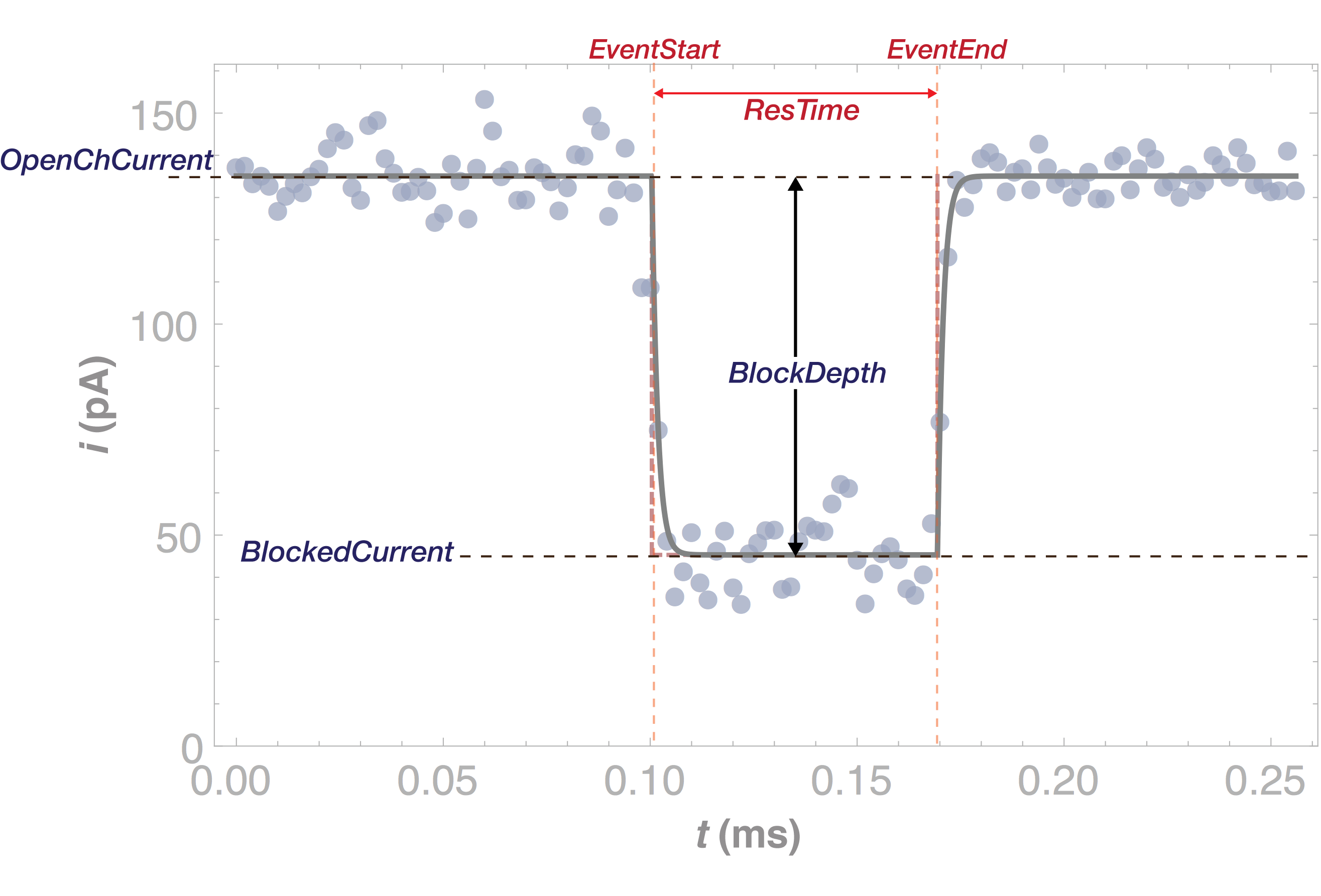
This simplification speeds up the analysis for two state events like the PEG event in the figure below. The figure shows the results of the fit (or meta-data) superimposed on the time-series of a single event.
Algorithm Settings¶
Metadata Output¶
Meta-data for individual events generated by adept2State can be queried using SQLite as described in the Database Structure and Query Syntax section. A list of meta-data stored by the step response algorithm is given below.
Column Name |
Column Type |
Description |
|---|---|---|
recIDX ProcessingStatus OpenChCurrent BlockedCurrent EventStart EventEnd BlockDepth ResTime RCConstant1 RCConstant2 AbsEventStart ReducedChiSquared ProcessTime TimeSeries |
INTEGER TEXT REAL REAL REAL REAL REAL REAL REAL REAL REAL REAL REAL REAL_LIST |
Record index. Status of the analysis. Open channel current in pA. Blocked state current in pA. Event start in ms. Event end in ms. BlockedCurrent/OpenChCurrent. EventEnd-EventStart in ms. Downstroke RC constant in ms. Upstroke RC constant in ms. Global event start time in ms. Reduced Chi-squared of fit. Event processing time in ms. (OPTIONAL) Event time-series. |