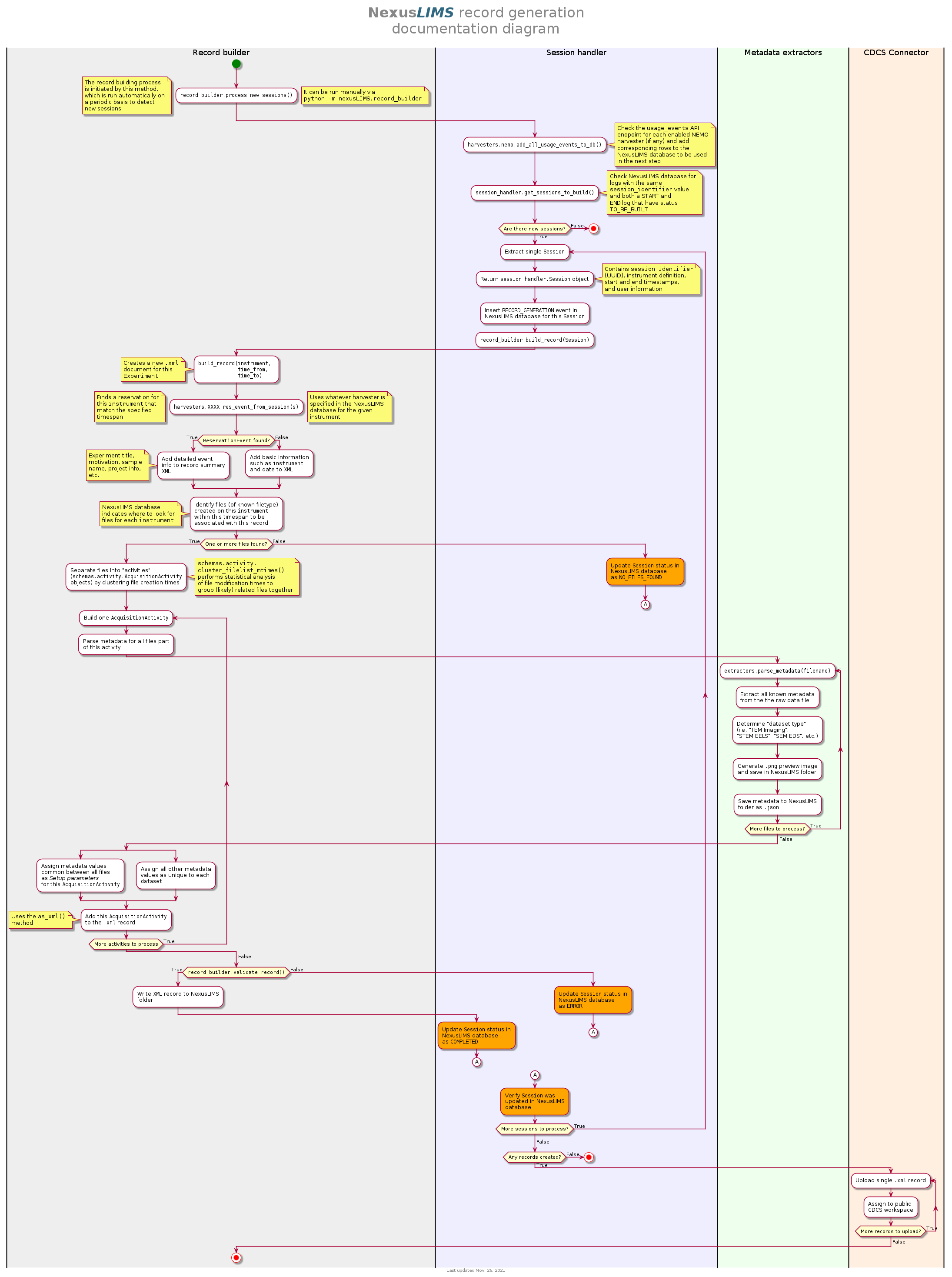
Record building workflow¶
Last updated: November 26, 2021
This page describes the process used to build records based on the data saved by instruments in the Electron Microscopy Nexus facility using NexusLIMS. At the bottom is an activity diagram that illustrates how the different modules work together to generate records from the centralized file storage and the NexusLIMS session database. Throughout this page, links are made to the API Documentation as appropriate for further detail about the methods and classes used during record generation.
General Approach¶
Because the instruments cannot communicate directly with the NexusLIMS back-end,
the system utilizes a polling approach to detect when a new record should be
built. The process described on this page happens periodically (using a system
scheduling tool such as systemd or cron). The record builder is begun by
running python -m nexusLIMS.builder.record_builder, which will run the
process_new_records() function. This
function initiates one iteration of the record builder, which will query the
NexusLIMS database for any sessions that are waiting to have their record built
and then proceed to build them and upload them to the NexusLIMS CDCS instance.
As of November 2021, it also queries the API for any enabled NEMO-based
harvesters, and will fetch any recent “usage events” to determine if a record
needs to be built.
As part of this process (and explained in detail below), the centralized file
system is searched for files matching the session logs in the database, which
then have their metadata extracted and are parsed into Acquisition Activities.
These activities are written to the .xml record, which is validated against the
Nexus Microscopy Schema, and finally uploaded to the NexusLIMS CDCS instance if
everything goes according to plan. If not, an error is logged to the database for that session
and the operators of NexusLIMS are notified so the issue can be corrected.
A note on authentication…
Since many of the resources accessed by the NexusLIMS back-end require
authentication (such as the SharePoint Calendar and the CDCS instance), it
is necessary to provide suitable credentials, or no information will be able
to be fetched. This is done by specifying two environment variables in the
context the code is run: nexusLIMS_user and
nexusLIMS_pass. The
values provided in these variables will be used for authentication to all
network resources that require it. The easiest way to do
this is by editing the .env.example file in the root of the NexusLIMS
repository and renaming it to .env (make sure not to push this file to
any remote source, since it has a password in it!). Furthermore, NEMO-based
session harvesting is enabled by specifying one or more sets of environment
variables named NEMO_address_X and NEMO_token_X, where X is an
integer value (1, 2, 3, etc.). The address variable should be
a path to the API endpoint for a particular server (e.g.
https://www.nemo.com/api/), while the token should be an API
authentication token created on the “Detailed administration” page of the
NEMO administration tools (ask your NEMO system administrator for more
direction if this is unclear).
Finding New Sessions¶
The session finding is initiated by
process_new_records(). This method
uses add_all_usage_events_to_db() to first
query any enabled NEMO API Connectors, and adds rows to the NexusLIMS session
log database for any newly created usage events (default is to look back seven
days for usage events). The record builder then calls
build_new_session_records(), which in
turn uses get_sessions_to_build() to
query the NexusLIMS database for any sessions awaiting processing (the database
location can be referenced within the code using the environment variable
nexusLIMS_db_path (e.g.
os.environ["nexusLIMS_db_path"]. This method interrogates the database for
session logs with a status of TO_BE_BUILT using the SQL query:
SELECT (session_identifier, instrument, timestamp, event_type, user)
FROM session_log WHERE record_status == 'TO_BE_BUILT';
The results of this query are stored as
SessionLog objects, which are then
combined into Session objects by
finding START and END logs with the same session_identifier value.
Each Session has five attributes
that are used when building a record:
- session_identifier
strAn identifier for an individual session on an instrument. This is a UUIDv4 for any Sharepoint Calendar events, but will be a resolvable URL for any NEMO-based events
- instrument
InstrumentAn object representing the instrument associated with this session
- dt_from
datetimeA
datetimeobject representing the start of this session- dt_to
datetimeA
datetimeobject representing the end of this session- user
strThe username associated with this session (may not be trustworthy, since not every instrument requires the user to login)
The get_sessions_to_build() method
returns a list of these Session
objects to the record builder, which are processed one at a time.
Building a Single Record¶
With the list of Session instances
returned by get_sessions_to_build(), the
code then loops through each Session,
executing a number of steps at each iteration (which are expanded upon below —
the link after each number will bring you directly to the details for that
step).
Overview¶
(link) Execute
build_record()for theInstrumentand time range specified by theSession
(link) Fetch any associated calendar information for this
Sessionusing one of the enabled harvesters (currentlysharepoint_calendarornemo).(link) Identify files that NexusLIMS knows how to parse within the time range using
find_files_by_mtime(); if no files are found, mark the session asNO_FILES_FOUNDin the database usingupdate_session_status()and continue with step 1 for the nextSessionin the list.(link) Separate the files into discrete activities (represented by
AcquisitionActivityobjects) by inferring logical breaks in the file’s acquisition times usingcluster_filelist_mtimes().(link) For each file, add it to the appropriate activity using
add_file(), which in turn usesparse_metadata()to extract known metadata andthumbnail_generatorto generate a web-accessible preview image of the dataset. These files are saved within the directory contained in the nexusLIMS_path environment variable.(link) Once all the individual files have been processed, their metadata is inspected and any values that are common to all files are assigned as
AcquisitionActivitySetup Parameters, while unique values are left associated with the individual files.(link) After all activities are processed and exported to XML, the records are validated against the schema using
validate_record().(link) Any records created are uploaded to the NexusLIMS CDCS instance using
upload_record_files()and the NexusLIMS database is updated as needed.
1. Initiating the Build¶
Prior to calling build_record() for
a given Session,
insert_record_generation_event()
is called for the Session to insert a
log into the database that a record building attempt was made. This is done
to fully document all actions taken by NexusLIMS.
After this log is inserted into the database,
build_record() is called using the
Instrument and timestamps associated with
the given Session. The code
begins the record by writing basic XML header information before querying the
reservation system for additional information about the experiment.
(go to top)
2. Querying the Reservation Calendars¶
Since users must make reservations on either the SharePoint calendar or the
NEMO facility management system, these are
important sources of metadata for the experimental records created by NexusLIMS.
Information from these reservation “events” is included throughout the record,
although it primarily informs the information contained in the <summary>
element, including information such as who made the reservation, what the
experiment’s motivation was, what sample was examined, etc.
To obtain this information, the record builder uses whatever harvester module
is listed in the harvester column of the NexusLIMS database for that
instrument. (i.e. either nemo or
sharepoint_calendar as of version
1.1.0). Each of these modules implements a res_event_from_session method,
used by the record builder to return a
ReservationEvent object representing
the reservation that overlaps maximally with the unit of time (or a very simple
generic one, if no matching reservation was found). These functions operate
serve as an adaptor layer to allow NexusLIMS to generate structurally-uniform
representations of a reservation from differing calendaring applications.
From this point on, identical code is used regardless of the original source of
the reservation information. Once the
ReservationEvent is obtained,
it is serialized into XML format that is compatible with the Nexus Microscopy
Schema. (go to top)
3. Identifying Files to Include¶
The majority of the information included in an Experiment record is extracted
from the files identified as part of a given session on one of the Electron
Microscopy Nexus Facility microscopes. To do this, a few different sources of
information are combined. As described before, a
Session will provide an identifier,
the timespan of interest, as well as the
Instrument that was used for the
Session. The
Instrument objects attached to session logs
are read from the instruments table of the NexusLIMS database, and contain
known important information about the physical instrument, such as the
persistent identifier for the microscope, its location, the URL where its
reservations can be found, where it saves its files (relative to the directory
specified in the mmfnexus_path environment variable),
etc. Sourcing this information from the master database allows for one central
location for authoritative data. Thus, if something changes about the
instruments’ configuration, the data needs to be updated in one location only.
The following is an example of the information extracted from the database and
available to the NexusLIMS back-end software for a given instrument (in this
case the FEI Titan TEM in Building 223, connected to the SharePoint harvester):
Nexus Instrument: FEI-Titan-TEM-635816
API url: https://sharepoint.url.com/_vti_bin/ListData.svc/FEITitanTEMEvents
Calendar name: FEI Titan TEM
Calendar url: https://sharepoint.url.com/Lists/FEI%20Titan%20Events/calendar.aspx
Schema name: FEI Titan TEM
Location: ***REMOVED***
Property tag: 635816
Filestore path: ./Titan
Computer IP: ***REMOVED***
Computer name: TITAN12345678
Computer mount: M:/
Using the Filestore path information, NexusLIMS searches for files
modified within the Instrument’s path during
the specified timespan. This is first tried using the
gnu_find_files_by_mtime(), which attempts to use
the Unix find command by spawning a sub-process. This only works on Linux, and may
fail, so a slower pure-Python implementation (implemented in
find_files_by_mtime()) is used as a fallback if so.
All files within the Instrument’s root-level
folder are searched and only files with modification times with the timespan
of interest are returned. Currently, this process takes on the order of tens of
seconds for typical records (depending on how many files are in the instrument’s
folder) when using the gnu_find_files_by_mtime().
Basic testing has revealed the pure Python implementation of
find_files_by_mtime() to be approximately 3 times
slower.
If no files matching this session’s timespan are found (as could be the case if
a user accidentally started the logger application or did not generate any
data), the
update_session_status() method is
used to mark the session’s record status as 'NO_FILES_FOUND' in the
database, and the back-end proceeds with step 1 for
the next Session to be processed.
(go to top)
4. Separating Acquisition Activities¶
Once the list of files that should be associated with this record is obtained,
the next step is to separate those files into logical groupings to try and
approximate conceptual boundaries that occur during an experiment. In the
NexusLIMS schema, these groups are called AcquisitionActivities, which are
represented by AcquisitionActivity
objects by the NexusLIMS back-end.
To separate the list of files into groups, a statistical analysis of the file creation times is performed, as illustrated in Fig. 1 for an example experiment consisting of groups of EELS spectrum images. In (a), the difference in creation time (compared to the first file) for each file is plotted against the sequential file number. From this, it is clear that there are 13 individual groupings of files that belong together (the first two, then next three, three after that, and so on…). These groupings represent files that were collected near-simultaneously, and each group is a collection of files (EELS, HAADF signal, and overview image) from slightly different areas. In (b), a histogram of time differences between consecutive pairs of files, it is clear that the majority of files have a very short time difference, and the larger time differences represent the gaps between groups.
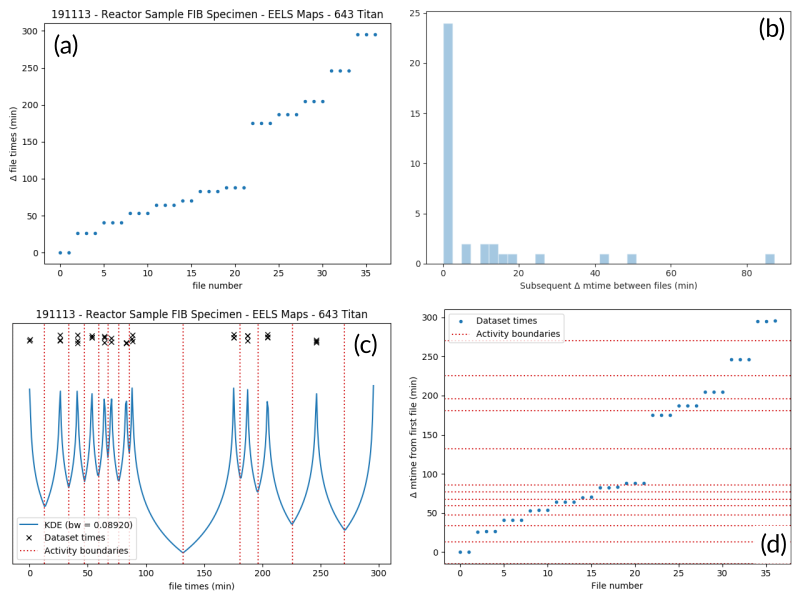
Fig. 1 An example of determining the
AcquisitionActivity time boundaries
for a group of files collected during an experiment. See the surrounding
text for a full explanation of these plots.¶
Since the pattern of file times will vary (greatly) between experiments, a
statistical approach is needed, as implemented in
cluster_filelist_mtimes(). In this method,
a Kernel Density Estimate (KDE) of the file creation times is generated. The
KDE will be peaked around times where many files are created in a short
succession, and minimized at long gaps between acquisition times. In practice,
there is an important parameter (the KDE bandwidth) that must be provided when
generating the density estimate, and a grid search cross-validation approach is
used to find the optimal value for each record’s files (see the documentation of
cluster_filelist_mtimes() for further
details). Once the KDE is generated, the local minima are detected, and taken
as the boundaries between groups of files, as illustrated in
Fig. 1 (c) (the KDE data is scaled for clarity).
With those boundaries overlaid over the original file time plot as in Fig. 1 (d), it can be seen that the method clearly delineates between the groups of files, and identifies 13 different groups, as a user performing the clustering manually would, as well. This approach has proven to be generalizable to many different sets of files and is robust across filetypes, as well. (go to top)
5. Parsing Individual Files’ Metadata¶
Once the files have been assigned to specific
AcquisitionActivity objects, the
instrument- and filetype-specific metadata extractors take over. These are all
accessed by the single parse_metadata() function,
which is responsible for figuring out which specific extractor should be used
for the provided file. The extractors are contained in the
nexusLIMS.extractors subpackage. Each extractor returns a
dict, containing all known metadata in its native (or close to)
structure, that has a top-level key 'nx_meta' containing a dict
of metadata that gets fed into the eventual XML record (note, this is not
currently enforced by any sort of schema validation, but will hopefully be in
the future). In general, the 'nx_meta' dict can be of arbitrary
depth, although any nested structure is flattened into a dict of
depth one with spaces separating nested keys, so it is important to avoid
collisions. Apart from a few special keys, the key-value pairs from the
'nx_meta' dict are reproduced verbatim in the XML record as
either Setup Parameters or Dataset Metadata, and will be displayed in the
CDCS front-end alongside the appropriate <AcquisitionActivity> or
<dataset>. Again, these values are not subject to any particular schema,
although this would be good place for validation against an instrument- or
methodology-specific ontology/schema, were one to exist.
As much as possible, the metadata extractors make use of widely adopted third-party libraries for proprietary data access. For most data files, this means the HyperSpy library is used, since it provides readers for a wide variety of formats commonly generated by electron microscopes. Otherwise, if a new format is to be supported, it will require decoding the binary format and implementing the extractors/preview generator manually.
parse_metadata() will (by default) write a JSON
representation of the metadata it extracts to a sub-directory within the
directory contained in the nexusLIMS_path environment
variable that matches where the original raw
data file was found in the directory from the
mmfnexus_path environment variable. A link to
this file is included in the outputted XML record to provide users with an easy
way to query the metadata for their files in a text-based format. Likewise, the
parse_metadata() function also handles
generating a PNG format preview image, which is saved in the same folder as the
JSON file described above. The actual preview generation is currently
implemented in
sig_to_thumbnail() for files
that have a HyperSpy reader implemented, and in
down_sample_image() for
simpler formats, such as the TIF images produced by certain SEMs.
The metadata dictionaries and path to the preview image are maintained at the
AcquisitionActivity level for all the
files contained within a given activity. (go to top)
6. Determining Setup Parameters¶
For each AcquisitionActivity, the
record builder will identify metadata keys/values that are common across all the
datasets contained in the activity after the individual files have been
processed, and stores these values at the <AcquisitionActivity> level of the
resulting XML record rather than at the <dataset> level. This allows for
easier determination in the front-end of what metadata is unique to each file
and also to see what metadata does not change during a given portion of an
experiment.
The code to do this determination is implemented in
store_setup_params(),
which loops through the metadata of each file of the given
AcquisitionActivity, testing to see if
the values are identical in each file. If so, the metadata value is stored as an
Activity Setup Parameter.
Once this process has completed,
store_unique_metadata()
compares the metadata for each file to that of the
AcquisitionActivity, and stores only
the values unique to that dataset (or at least not identical among all files
in the AcquisitionActivity).
(go to top)
7. Validating the Built Records¶
After the processing of each
AcquisitionActivity is finished, it is
added to the XML record by converting the Python object to an XML string
representation using
as_xml(). Once this has
been done for all the activities identified in the
earlier steps, the record is completed.
It is returned (as a str) to the
build_new_session_records() function,
and is validated against the NexusLIMS schema using
validate_record().
If the record does not validate, something has gone wrong and an error is
logged. Correspondingly, the
update_session_status() method is
used to mark the session’s record status as 'ERROR' in the database so the
root cause of the problem can be investigated by the NexusLIMS operations team.
If the record does validate, it is written to a subdirectory of nexusLIMS_path (environment variable) for storage before it is uploaded to the CDCS instance.
Regardless, the back-end then proceeds with step 1
for the next Session to be processed,
and repeats until all sessions have been analyzed.
(go to top)
8. Uploading Completed Records and Updating Database¶
Once all the new sessions have been processed, if there were any XML records
generated, they are uploaded using the
upload_record_files() function of the
nexusLIMS.cdcs module. This function takes a list of XML files to
upload, and attempts to insert them in the NexusLIMS CDCS instance using the
REST API provided by CDCS (documented
here).
The CDCS instance will validate the record again against the pre-loaded
NexusLIMS schema. upload_record_files() then assigns
the record to the Global Public Workspace so it is viewable without login.
Note: this will be changed in future versions once single-sign-on is
implemented, since records will be owned by the user that creates them.
At this point, the record generation process has completed. This entire logic is looped periodically as described at the top to continually parse new sessions, as they occur. (go to top)
Record Generation Diagram¶
The following diagram illustrates the logic (described above) that is used to
generate Experiment records and upload them to the NexusLIMS CDCS instance.
To better inspect the diagram, click the image to open just the figure in
your browser to be able to zoom and pan.
The diagram should be fairly self-explanatory, but in general: the green dot
represents the start of the record builder code, and any red dots represent a
possible ending point (depending on the conditions found during operation). The
different columns represent the parts of the process that occur in different
modules/sub-packages within the nexusLIMS package. In general, the diagram
can be read by simply following the arrows. The only exception is for the orange
boxes, which indicate a jump to the other orange box in the bottom left,
representing when an individual session is updated in the database.