Instructions
Installation
At the moment, this toolkit is only available outside of NIST through GitHub (the preference, either by fork, clone, or download) or directly from one of this book’s authors. For now, this toolkit includes the NIST PFAS Spectral Library. It is best used as an R project which can be opened directly in the RStudio Integrated Development Environment (IDE)2 which may be downloaded and installed free of charge if not already installed on a target system. Initial set up does require an internet connection to download software installers and dependencies; on a system which does not contain any software components this can take a considerable amount of time.
System Requirements
DIMSpec has been tested on both Windows 10 and Ubuntu 20.0.4.3 LTS 64-bit3 platforms and should run on any system able to install R, Python, SQLite3, and a web browser, though installation details may vary for other operating systems. Follow the instructions for each requirement on the target operating system.
[REQUIRED] R 4.1+ (download) and many packages are required (R Core Team (2021); various); necessary packages will be installed when the compliance file is sourced, which may take some time when the project is first installed. The RStudio IDE (download; RStudio Team (2020)) is highly recommended for ease of use as this project is distributed as an R project.
[STRONGLY RECOMMENDED] SQLite3 (download) and its command line interface (CLI; download) provide the database engine in structured query language (SQL) and are not technically required as the build can be accomplished purely through R, but are highly recommended to streamline the process and manipulate the database. A lightweight database interface such as DBeaver Lite is also suggested for interacting with the database in a classical sense. Git (install instructions) is a repository manager which will make it much easier to install and update the project. The sqlite3 CLI and git executables must be available via PATH.
[RECOMMENDED] For chemical informatics support, both Python 3.9+ and the rdkit (RDKit: Open-Soure Cheminformatics (version 2021.09.4), n.d.) library are required for certain operations supporting display and calculations, primarily generation of machine-readable identifiers (e.g. InChI, InChIKey, SMILES, etc) but the full capabilities of rdkit are available (see the RDKit documentation for details); these are turned on by default but are completely optional. An anaconda or miniconda installation is required. Python integration is not required for spinning up the basic database infrastructure. Users may need to add the conda executable to their PATH and, if conda is already installed, should pay close attention to the Python section of Technical Details. If these are not available, R will install miniconda (this requires user confirmation at the console) and create the necessary environment as part of automated setup during the compliance script. (Another option for chemical informatics is to use the Java-based R package rcdk instead; users will need to install the Java framework prior to installing rcdk (see Windows; Ubuntu). This package is not well supported in this project and rdkit is preferred.)
[OPTIONAL] It is helpful to have some data on hand to populate and evaluate the database. Every effort has been made to simplify the process of building databases using this tool, and data can be populated from CSV files of a defined structure; examples are provided but the process of generating them can be somewhat onerous as key relationships must be defined to automatically populate in this manner. Future work may be able to simplify this process further, if necessary, but for now, interested researchers are encouraged to contact the authors for guidance on how to transform data to fit this schema.
The following sections provide more detailed information on how to use the tools provided to interact with the database and customize it for other uses.
Initial Setup
This section provides instructions in a “quick start” format. While every effort was made to make this as painless as possible, success may vary from system to system. This assumes that R v4.1 or later is installed. Several quick start guides offering more detail about aspects of the project, including installation, are also available in the repository (listed in the project README) and for download from the online User Guide by clicking the download icon in the header at the top of the User Guide to select your download of choice.
- If using RStudio:
- Open the project in RStudio.
- Open the file at
"R/compliance.R"in the editor. - Run the compliance script by clicking the “Source” button at the top right of the editor pane or typing
source("R/compliance.R")in the console pane.
- If not using RStudio:
- Open an R session in the project directory or launch R and set your working directory to that of the project (e.g.
setwd(file.path("path", "to", "dimspec_dir")). - Execute the command
source("R/compliance.R").
- Open an R session in the project directory or launch R and set your working directory to that of the project (e.g.
Using either method should in most cases establish the compute environment, activate logging and argument validation, bind to a python environment providing rdkit support, launch an API server, and list out the web applications available. The project is distributed with a database populated with high resolution mass spectrometry data for per- and polyfluoroalkyl substances (PFAS) for evaluation purposes both to distribute this data set and to evaluate capabilities for reuse in other projects.
Project Directory
The project directory contains the following directories of interest. Generally, the only modifications (if any) that should be necessary to operate DIMSpec projects are to environment establishment files located in the /config directory or in the /inst directory if extending the DIMSpec API or web applications.
/configFiles pertaining to the build, description, and population of the underlying database, as well as certain compute environment settings and import settings. This serves for rapid rebuilding and reuse of the underlying database structure. Also included are environment establishment files for the project (“env_glob.txt”), the R session (“env_R.R”) and the optional logging (“env_logger.R”) functionality./sql_nodesFiles containing the sql scripts defining the database schema, as run by the “build.sql” script. Files are separated into database “nodes” with the hope that many can be repurposed or used a la carte in future projects. A graphical representation of the database schema, the entity-relationship diagram (ERD) is also available./dataComma-separated-value (CSV) files which can be used to populate tables defined by their SQL nodes and which will be populated according to the chosen population script. This directory contains common data which should be applicable to all database produced by this tool (i.e. normalization tables, elements and isotopes, etc.) and subdirectories containing project-specific CSV files.
/exampleFiles providing examples of (mainly) import files in JavaScript Object Notation (JSON) format. These are the files used to populate empirical data and were produced by the NIST Non-Targeted Analysis Method Reporting Tool./imagesIf images of molecular models are produced using rdkit through this toolset, they will be housed here, named by the molecule’s known structure identifier (e.g. SMILES, InChI, etc.). Other images may be produced during routine work and should also be placed in this directory, though user-produced images and graphics can be saved anywhere./instFiles for rdkit integration (/rdkit), the API service (/plumber), and Shiny applications (/apps)./rdkitEnvironment establishment and rdkit functions are located here. These will determine how R connects to the python environment to integrate rdkit into an R session as well as the files necessary to build the environment (e.g. “environment.yml”). Functions in the “py_setup.R” file should suffice for most use cases./plumberEnvironment establishment and API definition functions are located here. These will determine how requests to the API are routed and functionality are provided through http protocols in a RESTful manner. It comes complete with Swagger documentation available when the server is running./appsEnvironment establishment, general resources, and shiny application files are located here. Each application is contained within its own directory.
/logsIf logging functionality is turned on, logs will be written here according to the namespace of the log (e.g. logs written to the “db” namespace will be written to"logs/log_db.txt")./Rdirectory; most general R functions are housed here or in one of the subdirectories.
Project Set Up
Running the compliance script at "/R/compliance.R" will establish the project for you in most cases. It leverages several files to determine project settings; these are detailed here for clarity and customization options, with further details provided in the Compute Environments section. To accept the default settings, source the compliance file and move on to the Using DIMSpec section. This may take a while to resolve package dependencies. To customize your implementation, read on. Changing any of these settings is entirely optional.
Step 1 - Customize global environment settings
Several options are available to customize the use of DIMSpec to any given project component; settings are in the file "/config/env_glob.txt". These values are not set at the system level to add flexibility across operating systems; they are instead session values that are available while a session is active.
| Setting | Type | Description |
|---|---|---|
| DB_TITLE | String | The title to use for this implementation. |
| DB_NAME | String | The name of the database to create or use. For SQLite this should be the name of the database file. |
| EXPLICIT_PATHS | Logical | Whether or not file names are fully qualified with their path. |
| DB_BUILD_FILE | String | The .sql file name of the script used to build the database (e.g. “build.sql”; see Database Schema. |
| DB_BUILD_FULL | String | The .sql file name of the fallback build script that should be used if the sqlite3 command line interface (CLI) tool is not available (e.g. “build_full.sql”; see Database Schema). |
| DB_DATA | String | The .sql file name of the data population script to run when populating the database at build time (e.g. “populate_common.sql”; see Populating Data). |
| SQLITE_CLI | String | The name of the terminal command to launch the sqlite shell (e.g. sqlite3). This must be available in your PATH. |
| CONDA_CLI | String | The name of the terminal command to execute ana-/miniconda commands (e.g. conda). This must be available in your PATH. |
| INIT_CONNECT | Logical | Whether or not to connect to the database when starting a session by sourcing the compliance script. |
| LOGGING_ON | Logical | Whether or not to establish an environment to perform action logging, which will carry additional information about what functions in the DIMSpec toolkit are doing (see Logger). |
| USE_API | Logical | Whether or not to activate the plumber application programming interface (API) for this session (see Plumber). If this is set to TRUE, the plumber service will launch in a background process by default and return control to the console. |
| API_LOCALHOST | Logical | Whether or not to activate the plumber application programming interface (API) hosted on the local machine only. If this is set to FALSE, the API will be hosted and available on your network. |
| API_HOST | String | If API_LOCALHOST is set to FALSE, set this to the IP address or network path to the computer hosting the API. |
| API_PORT | Integer | The hosting port on which to launch the plumber service. This must be an open port or the launch will fail. If API_LOCALHOST is TRUE, the API will launch at “localhost:API_PORT”, otherwise at “API_HOST:API_PORT”. |
| INFORMATICS | Logical | Whether or not to establish an environment providing informatics support, primarily with RDKit. To streamline installation of only the database and R tools, set this to FALSE. |
| USE_RDKIT | Logical | Whether or not to use RDKit for informatics (requires python). If set to FALSE, the packages BiocManager, ChemmineR, and rcdk will be installed if not available, though support for these is not provided at this time. |
| USE_SHINY | Logical | Whether or not to establish an environment providing support for web applications provided as part of the project (defaults to TRUE). |
| SHINY_BG | Logical | [PLANNED FEATURE] Whether or not to launch shiny apps as part of a background process, making them immediately available from a web browser when the compliance script is executed (defaults to FALSE). |
Step 2 - Customize R session settings in the “env_R.R” file
More customization options that require R are available to set up the project specifically for your application. Open the file "config/env_R.R" to customize these for your use. These values are not set at the system level to add flexibility across systems; they are instead session values that are available during use of the project, and many depend on settings from the section above, which will be applied automatically if they are not already set.
| Setting | Type | Description |
|---|---|---|
| DB_DATE | Date | The date the database file was last created, as determined by file properties. Override with a date value (e.g. as.Date("2022-06-01")) |
| DB_RELEASE | String | The major and minor release versions for this database. |
| DB_VERSION | Generated String | Combines the DB_RELEASE and DB_DATE (if built) values for a complete version of the database. |
| DB_PACKAGE | String | The name of the R package allowing connection to your database (e.g. RSQLite in most cases, but could be any database connection package). |
| DB_DRIVER | String | The name of the database driver function allowing connection to your database, which must be a function available in DB_PACKAGE (e.g. SQLite). |
| DB_CLASS | String | The class of an R object resulting from a call to the function defined by DB_PACKAGE::DB_DRIVER (e.g. SQLite); this will be used to search for and manage connections in the session. |
| DB_CONN_NAME | String | The name to be used for the R object database connection (e.g. con in most cases here); this defaults to a session variable named DB_CONN_NAME if it exists to facilitate independent management of multiple connections. |
| DEPENDS_ON | String Vector | The list of packages required by your project. The list provided is the bare minimum required for functionality in the project as distributed. Add more to expand functionality for your use cases if necessary. |
| EXCLUSIONS | String Vector | The list of files and directories to exclude from automatic loading when the compliance script is run. |
| IMPORT_MAP | String | Imports the mapping file determining relationships between import files and the database structure (see Importing Data); change the name of the CSV file (change also the function calling it if using formats other than CSV) to point to a different map. |
| LOGGING_ON | Logical | Whether to activate logging functionality when a session begins. This defaults to the session variable named LOGGING_ON and, if not present, to TRUE. If TRUE, adds the logger package to the dependency list. |
| VERIFY_ARGUMENTS | Logical | Whether or not to activate function argument verification for this project. The default of TRUE will check arguments provided to many functions for compliance with function expectations and is good for development work, but also slows down execution times. Set to FALSE to turn this off. |
| MINIMIZE | Logical | If TRUE, turns off both LOGGING_ON and VERIFY_ARGUMENTS to speed up execution time. |
| USE_API | Logical | Defaults to the global setting of USE_API. If TRUE, several options are provided to customize properties of the API. Set these as appropriate for advanced use cases; the defaults will make the API available on your local system at http://127.0.0.1:8080. |
| USE_SHINY | Logical | Defaults to the global setting of USE_SHINY. If TRUE, a list of installed shiny applications will be available to your session under the named character variable SHINY_APPS which contains absolute paths to the application directories. These are launchable (and should resolve their environment) at any time during a session from the console using shiny::runApp(SHINY_APPS["app_name"]) where "app_name" is the name of a shiny app in the variable. See the Mass Spectral Match for Non-Targeted Analysis (MSMatch) application that ships with this project as an example to match user supplied mass spectral data against the library. |
Step 3 - Customize logger settings in the “env_logger.R” file
To provide support information about performance and support troubleshooting, a logging utility is provided with the project. Logs are managed by namespace and generated with the function log_it which uses the logger package for additional functionality (see Logger). Customization options for the format of these logging messages are provided, though under most circumstances should be left as-is to support reading logs back into a session. These values are not set at the system level to add flexibility across systems; they are instead session values that are available during use of the project, and many depend on settings from the sections above, which will be applied automatically if they are not already set.
Three support functions are also provided in this file to update the logger settings during a session (update_logger_settings), read logs from a file back into the session (read_log), and convert a log file into a session data frame for deeper inspection (log_as_dataframe).
Environment set up files that follow the same approach are also provided for rdkit integration, the plumber API server, and shiny web applications; these are not detailed here and should only be changed when necessary. See those sections in Technical Details for more information.
| Setting | Type | Description |
|---|---|---|
| LOG_DIRECTORY | String Path | The relative path to the project directory housing logs. This defaults to the session variable named LOG_DIRECTORY and, if not present, to "logs". If that directory is not present, it will be created. |
| layout_console | Function String | The format to use when printing logs to the console, by default interpreted by logger::layout_glue_generator, but could be any logger generator. |
| layout_file | Function String | The format to use when printing logs to a file, by default interpreted by logger::layout_glue_generator, but could be any logger generator. |
| log_remove_color | Regex String | A regular expression describing color formatting to strip out when reading logs back into a data frame. If printing to the console in RStudio, colors will be maintained. This should coordinate with the layout_console format. |
| log_split_column | Regex String | A regular expression describing the character formatting used to split log records into columns when reading logs back in as a data frame object with the log_as_dataframe function. This should always be coordinated with the layout_file format. |
| LOGGING | List | A nested list object defining logging settings for different namespaces. Each must include the following named settings:
New namespaces can be added during the session if desired, but this list should define the most common ones. More information about the logging environment is provided in the Logger section of Technical Details. |
| LOGGING_WARNS | Logical | Whether to log all warning messages generated during this session by default. |
| LOGGING_ERRORS | Logical | Whether to log all error messages generated during this session by default. |
Using DIMSpec
There are several R packages required for this project, so initial set up may take some time. To streamline this process once set up is complete, a compliance script is available that will install and load required packages; run source("R/compliance.R") in the console to establish the runtime environment. See References for the complete list of library dependencies. Based on project settings, components can be turned on or off as desired for lighter weight applications. In many cases helper functions are available to turn these components back on during an active session without interrupting the current environment. The following sections assume the compliance script has run and that all functions are available. At any time, use fn_guide() or fn_help("fn") where "fn" is the name of a function (quoted or unquoted) to view function documentation from within R.
Database Connections
Connecting to an Existing Database
This project uses SQLite by default as a portable database engine where the database is contained to a single file. To connect a project to a particular database (e.g. you have multiple databases for different projects), simply change the value of DB_NAME in “env_glob.txt” prior to sourcing the compliance file. The database distributed with the project contains mass spectral data for per- and polyfluoroalkyl substances as an example. It (and any databases created using this project), opens in write-ahead logging (WAL) mode for speed and concurrency. This does generally require the database file to be present on the same machine as the project but allows installation on instrument controllers that may not comply with network security restrictions. As with all SQLite databases, foreign key enforcement must be turned on when connecting with pragma foreign_keys = on; the manage_connection function takes care of this and other connection management aspects automatically and is the recommended way to connect and disconnect to DIMSpec databases. Call manage_connection(reconnect = FALSE) to close the connection. Calling manage_connection calls DBI::dbConnect and DBI::dbDisconnect with certain checks and parameter defined side effects to manage the connection.
Creating a New Database
Tooling to create a new SQLite database using this schema is built into the project; functions are in the “R/db_comm.R” file and help documentation is available from within the project using the fn_guide and fn_help functions. When creating a new database, prior to sourcing the compliance file, set options in the “env_glob.txt” and “env_r.R” files appropriately. If the file identified by DB_NAME does not exist it will be created according to the SQL scripts selected as DB_BUILD_FILE and DB_DATA; edit those files if necessary for your use case. To build a new, empty database users need only set DB_NAME to a file that does not exist in the project directory, and DB_DATA to “populate_common.sql” which contains the majority of source data necessary to populate normalization tables (see the Database Schema and Populating Data sections for more detail).
Alternatively, once the compliance file has been sourced, a new database may be created directly from R with the build_db function; this function takes as default values those provided in the environment, but you can at any time define different specifications. For example, to create a new database with a different SQL definition and population script use:
build_db(
db = “new_database.sqlite”,
build_from = “this_file.sql”,
populate = TRUE,
populate_with = “new_data.sql”,
connect = FALSE
)If a connection already exists that you wish to maintain in the session, be sure to call this with connect = FALSE in order to not drop the connection (see next section for managing multiple connections). If you do not wish to maintain a connection to the previous database, this can be safely called with connect = TRUE (the default) and the prior connection will be replaced with the new one.
Connecting to Multiple Databases
If your project needs to connect to multiple databases, separate connections can be made and managed within a single R session. For convenience, the supplied manage_connection function will apply to the database and connection object defined in the setup files (see Project Set Up). Enable new connections alongside existing connections (e.g. the one created in the previous section) with manage_connection(db = “new_database.sqlite”, conn_name = “con2”) where db points to the new database file and conn_name does not exist in the current environment. There is no limit to the number of connections that can be made in this manner, and the WAL will be flushed each time this function is called if no other connections exist.
Using a Database Connection in an R Session
If INIT_CONNECT = TRUE, sourcing the compliance file will establish a connection to the database named in DB_NAME and make the connection available as an R session object with the name defined by DB_CONN_NAME (the default is con). Several convenience functions are available with those options set.
Functions from the dplyr package support database operations as implemented in the dbplyr package, meaning you can work with database objects using the “tidyverse” as if they were local objects (e.g. tbl(src = con, “contributors”) where con is your database connection object and “contributors” is the name of a database table or view). Simple database operations (e.g. filters, joins, column selection, etc) are supported and the resulting object is an external pointer to a lazy database query; to pull data as a data frame (e.g. necessary to join a local data frame with a database query result) use collect() on the tbl object. There are, however, some tasks (e.g. complicated or programmatic queries) where that may prove insufficient. In that case, two options are available.
The connection object fully supports direct communication for SQL queries through the DBI package and is likely a familiar option for users comfortable with SQL. To continue the example, dbGetQuery(con, “select * from contributors”) will return the same data as in the tbl example above, except that it returns a data frame rather than a pointer object.
For users less familiar with SQL, the function build_db_action is provided to support nearly all database operations. There may be edge cases where it fails. Results from the following function are equivalent to the dbGetQuery result but will construct the query programmatically, allowing for the passing of arguments and always returning a data frame:
build_db_action(
action = “select”,
table_name = “contributors”
)As this function performs argument verification and SQL interpolation to protect queries from unintended side effects, this is the recommended manner to directly interact with the database for anything other than basic queries. It supports typical database actions (including SELECT, INSERT, UPDATE, and DELETE, as well as a custom GET_ID action that returns an integer vector of the id column for all records matching the query) and operations (GROUP BY, ORDER BY, DISTINCT, LIMIT). Search and filter options can be passed programmatically to match_criteria as a list and are parsed by the clause_where function.
Queries do not have to be executed; set the argument execute = FALSE to examine queries prior to execution or save common queries for reuse. See the full function reference with for advanced use of the build_db_action and clause_where functions with fn_help.
Inspecting Database Properties
Code decoration conventions used in the SQL files enable reading table definitions and properties from SQLite into R with the function pragma_table_info. Supply the name of a database table or view to get information about that table; different connections can also be used for comparison if desired. This is the interactive version; a version in JSON format can be saved using save_data_dictionary. This saved file is loaded during the compliance script as object db_dict which is a named list of data frames; names correspond to database entities. This can be regenerated and brought back into the R session at any time (see data_dictionary) and should be updated if modifications are made to the underlying schema.
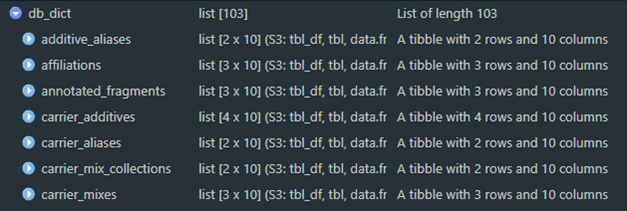
Figure 1a. An example of the data dictionary object.
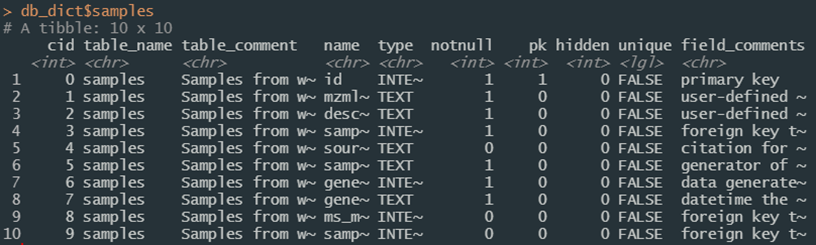
Figure 1b. Details of the “samples” table from the data dictionary.
Relationships between database entities can also be queried programmatically. Use the er_map function to read the same decoration convention in the SQL definitions to extract relationships. An object is created during the compliance script as db_map to make it available to your session. This results in a nested list with names corresponding to database entities, and elements describing the object name, its type, which table(s) and column(s) it references, which table(s) reference it, which table(s) it normalizes, and which view(s) use it.
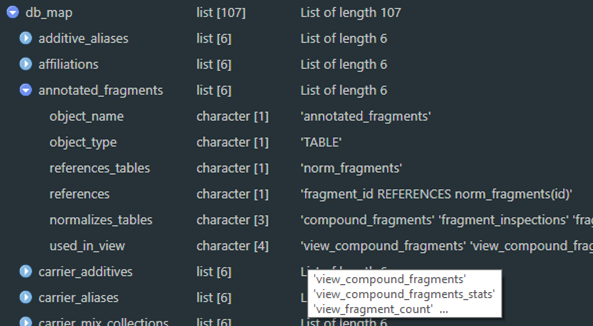
Figure 2a. An example of the R object structure of an entity map as a list.
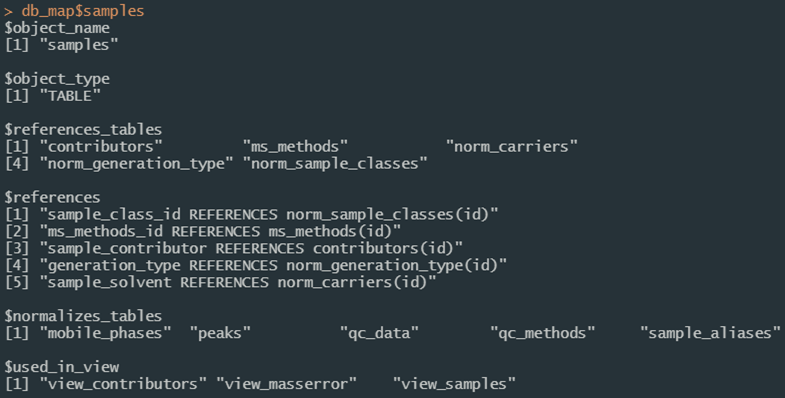
Figure 2b. Details of the “samples” table from the data entity map object in Figure 2a.
Using the Application Programming Interface (API)
Application Programming Interfaces (APIs) enable software components to communicate with each other. Most modern machine communication happens through APIs. In the context of this project, an API server is launched using the plumber package to reduce computational load on R sessions or shiny applications and ensure consistent results across multiple sessions. It does not have to be used (set USE_API = FALSE in “env_glob.txt” to turn it off) but is encouraged and is a requirement for all shiny applications that ship with this project.
The compliance script launches this in a background process by default at http://localhost:8080. Use api_open_doc to open the documentation page directly in a browser. To start the service manually from an interactive session and load the documentation immediately for exploration and testing, use api_reload(background = FALSE); if it is already running in a background process and desirable to launch a second service (e.g. for testing new endpoints or changes to existing ones), set the pr parameter to a different name and the on_port parameter to an open port (it will fail if the port is already in use). Documentation is produced by Swagger and is interactive, allowing for users to enter values and get both the return and the URL necessary to execute that endpoint (Figure 3). See the Plumber section in Technical Details for more information. If the compliance script is run with USE_API = FALSE and api_reload is not available, it may be more intuitive to use start_api.
Endpoints for many predictable read and search interactions are available. Session variables define the connections, and communication and control functions default to those expected values for streamlining (e.g. functions like api_reload, api_open_doc, and api_endpoint may be called without referring explicitly to a session object or URL for the current project).
The main interactivity with the API from an R session or shiny application is through the api_endpoint function. The first argument (i.e. path) should always be the endpoint being requested. Additional named parameters are then passed to the API server; the same example endpoint result in Figure 3 called from the console would be
api_endpoint(
path = “compound_data”,
compound_id = 2627,
return_format = “data.frame”
)with an example of the results in Figure 4. Endpoints of most use to those using the service will vary according to needs and are detailed in the Plumber section in Technical Details. Call them with api_endpoint(path = *X*) and any other arguments required by the endpoint. Paths listed here are likely of most use:
- “_ping”, “db_active”, and “rdkit_active” indicate that the server is alive and able communicate with the database and rdkit, respectively;
- “list_tables” and “list_views” return available tables and views respectively;
- “compound_data” and “peak_data” return mass spectrometry data associated with a compound or peak and must be called with
compound_idorpeak_idequal to the database index of the request; in most cases these should be called withreturn_format = "data.frame"; - “table_search” is a generic database query endpoint analog for
build_db_actionto constructSELECTqueries and has the most parameters for flexibility; for more information seefn_help(build_db_action)for details; relevant parameters are summarized here:table_name should be the name of a single table or view;
column_names determine which columns are returned;
match_criteria should be a list of criteria for the search convertible between R lists and JSON as necessary; values should generally follow the convention
list(column_name = value)and can be nested for further refinement using e.g.list(column_name = list(value = search_value, exclude = TRUE))for an exclusion search (seefn_help(clause_where)for additional details); when called viaapi_endpointR objects can be passed programmatically;and_or should be either
"AND"or"OR"and determines whether multiple elements ofmatch_criteriashould be combined in an AND or OR context (e.g. whetherlist(column1 = 1, column2 = 2)should match both or either condition);limit is exactly as in the SQL context; leave as
NULLto return all results or provide a value coercible to an integer to give only that many results;distinct is exactly as in the SQL context and should be either TRUE or FALSE;
get_all_columns should be either
TRUEorFALSEand will ensure the return of all columns by overriding thecolumn_namesparameter;execute should be either
TRUEorFALSEand determines whether the constructed call results are returned (TRUE) or just the URL (FALSE); andsingle_column_as_vector should be either
TRUEorFALSEand, ifTRUE, returns an unnamed vector of results if only a single column is returned.
These and other endpoints can be easily defined, expanded, or refined as needed to meet project requirements. Use api_reload to refresh the server when definitions change, or test interactively prior to deployment using Swagger by launching a separate server either by opening the plumber file and clicking the “Run API” button in RStudio, or using the api_start or api_reload functions as described above. To support eventual network deployment, any number of API servers may be launched manually on predefined ports to allow for load balancing.
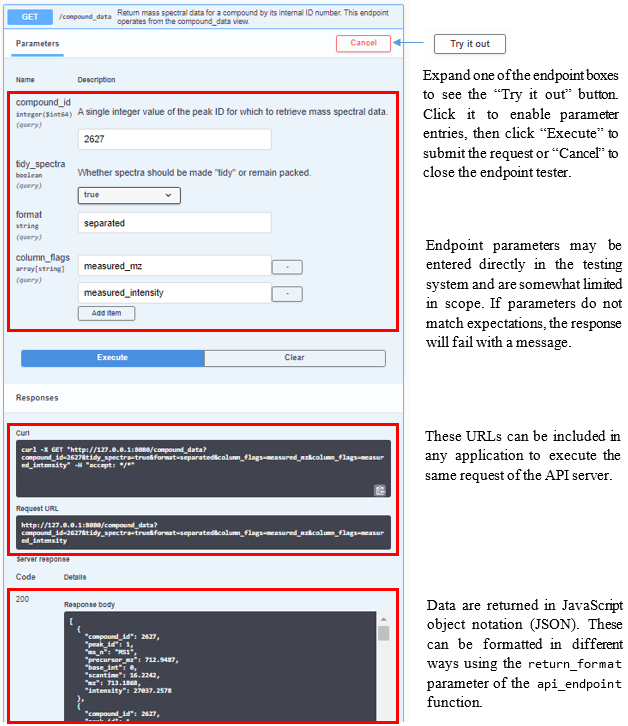
Figure 3. Screen shot and descriptions of the interactive Swagger documentation page for the endpoint /compound_data, available using api_open_doc(). Click the “Try It Out” button to activate the testing mode.
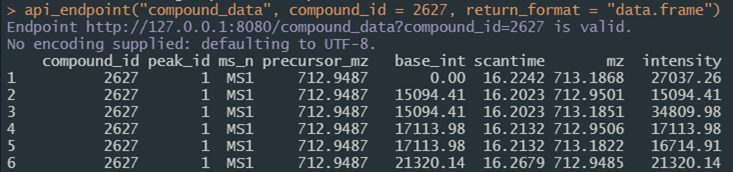
Figure 4. Screen shot of the result of calling the same API endpoint as in Figure 3 from an R session.
Using rdkit
For chemometrics integration, rdkit is made available as part of the project. This user guide does not provide details about rdkit; users are instead directed to the documentation for details. All functionality provided as part of rdkit is supported with some limitations through the reticulate package. In most cases the required environment should resolve during the compliance script. On certain systems it may be desirable to install the environment manually (instructions in the Python section of Technical Details).
Once an R session has activated and bound to a python environment it cannot be deactivated, but instead must be terminated to drop this binding. Once bound to a session object, all rdkit functions are accessible as a list of functions (just as in any python integration using reticulate) following rdkit module structures (e.g. rdk$Chem$MolFromSmiles("CN1C=NC2=C1C(=O)N(C(=O)N2C)C")). Though these can be chained together or piped, for stability it is recommended to store the return of each call as a variable; returned objects may not always be readily used in further functions.
A few custom R functions are made available to assist with the process. The implementation will depend on the environment definition found in “inst/rdkit/env_py.R” but in the standard use case will result in a session object named rdk tied to a python environment named “nist_hrms_db” using packages built from conda forge. See the function reference guide using fn_guide() for additional details, but the following functions are likely the most useful:
setup_rdkitis a convenience function that should install and bind to python in a session;rdkit_activeis the main check to determine whether or not rdkit has been bound to the current session and allows for setting multiple bindings if desired by settingrdkit_refto a different value, and will triggersetup_rdkitif called withmake_if_not = TRUE;molecule_picturecreates a graphic of a molecular model from structural notation and is an example ofrdkitfunctionality; andrdkit_mol_aliasesgenerates machine-readable structural notation in a variety of formats (e.g. InChI and InChIKey) given a notation with a known format and can interchange between these to create molecular aliases; all formats supported byrdkitare attempted ifget_aliases = NULL(Figure 5) but generally these would be specific by project needs; results that fail or are blank are removed and the return is by default a data frame to support any number of identifiers with one pass.
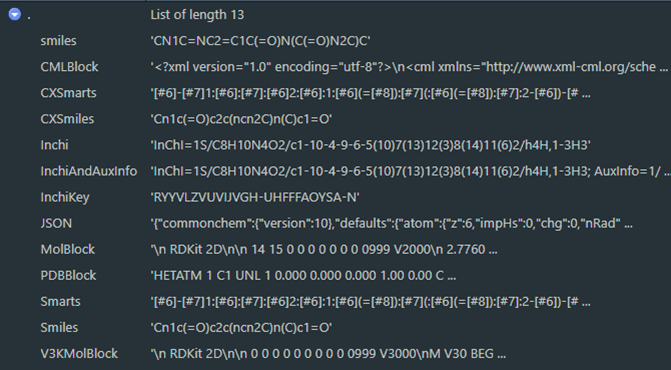
Figure 5. All molecular aliases as seen in the RStudio viewer for results of a call to rdkit_mol_aliases("CN1C=NC2=C1C(=O)N(C(=O)N2C)C", get_aliases = NULL)
Logging
Logging messages for statuses, information, warnings and errors are provided throughout functions used in this project and is executed through the log_it function. This function builds on top of the logger package to construct, decorate, and write to file any logging messages necessary, and offers console messages in case logger is unavailable. If logging is enabled and the logger package available, logs may also be written to files in the “logs” directory and later retrieved with the utility functions read_log and log_as_dataframe, whose first parameter is the name of the file to read from the /logs directory. Logs written to disk by default are separated by namespace (e.g. /logs/log_db.txt vs /logs/log_api.txt) to facilitate support, but output files may be defined as any available .txt file path and will be appended to existing files. Logs may look odd if viewed directly as they include text decorations to display in the console.
Settings are available for five namespaces by default (see Logger and Project Set Up for more details) as established by the “config/env_logger.R” file; more can be enabled at any time using the add_unknown_ns and clone_settings_from parameters of log_it. Logs can then be generated from within any function using e.g.:
log_it(
log_level = “info”,
msg = “Log message text”,
log_ns = “global”
)where log_level is the category of message, msg is the message itself, and log_ns is the namespace. Settings defined in the LOGGING session variable determine how logs are processed. Each message produced with log_it includes the timestamp, namespace, status (i.e. log_level), function calling the message, and the message itself. While log_it will print to the console messages of any level, log_level should be one of the supported logging levels (trace, debug, info, success, warn, error, or fatal) to integrate with logger, which is required if the logging message is to be written to a log file.
Users developing on top of this infrastructure are encouraged to take advantage of the logging functionality and make liberal use of the log_it function to ease debugging and maintenance.
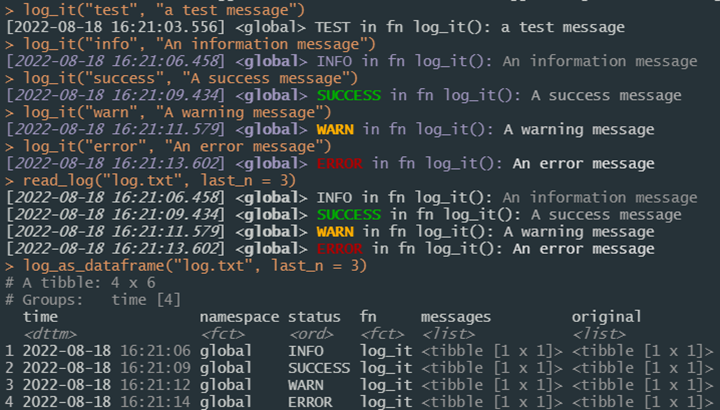
Figure 6. Example uses of log_it to create logging messages.
Using Shiny Applications
The Shiny package enables web applications written using R, which often meaningfully make custom processing code like that written for this project available to broader audiences. Additionally, inputs can easily be type verified and restricted to preset expectations. When the compliance script is run, a named vector of available shiny apps will be available as SHINY_APPS. These can be started with the start_app(app_name = X) where X is the name of the application as found in names(SHINY_APPS). Shiny apps are fluid and responsive; will automatically arrange themselves to best fit your browser size and can be custom designed with any layout or functionality. By default all communication with the database is routed through the plumber API.
This allows environment resolution to launch applications directly from the console, without any need to run the compliance script. Launching an app is then possible directly from the console (or batch file shortcuts which could be included in later updates) using e.g.
`shiny::runApp(“inst/apps/table_explorer”)`from the project directory.
Three shiny applications ship with this project as of the time this document was written.
table_explorerallows users to explore database tables and views by selecting it from a drop-down list and details definitions and connections to other tables and views; this app should be amenable to any database created with DIMSpec and is detailed in its own section;msmatchallows users to upload an mzML file of mass spectral data and search user-defined features of interest by mass to charge ratio and chromatographic retention time for matches in the database for both known compounds and annotated fragments, while providing contextual information about the method and samples used to generate reference spectra. The MSMatch application is detailed in its own section.dimspec-qcallows users to perform the quality control evaluation of potential imported data and generates the necessary JSON object to be incorporated into the database. The DIMSpec-QC application is detailed in its own section.
An application template is also included which should accelerate development of additional applications on top of the DIMSpec infrastructure to facilitate project needs.
Importing Data
For now, data imports are only supported from the command line using outputs generated by the NIST Non-Targeted Analysis Method Reporting Tool (NTA MRT). That tool is a macro-enabled Microsoft Excel® workbook available on GitHub that
“…allows for the controlled ontology of method data reporting and the export of the data into a single concise, human-readable file, written in a standard JavaScript Object Notation (JSON).”
Users fill out the workbook annotating features of interest and associated fragmentation identities. Generated method files are submitted alongside the mzML file (converted from instrumentation output using Proteowizard’s msConvert software (Adusumilli, Ravali and Mallick, Parag 2017). After quality control checks are performed, the resulting JSON object holds everything necessary to import data into the database.
Data passing quality control checks (see the DIMSpec Quality Control section for a shiny application to check quality control aspects of mzML files) are imported using functions found primarily in the /R/NIST_import_routines.R file. Field mapping is defined by the /config/map_NTA_MRT.csv file, which contains a list of import file elements and their properties, with connections for each to their destination tables and columns; individual elements are resolved by the map_import function which does much of the transformation. New maps can be created and used in support of other import formats in the future, and as the import functions are heavily parameterized they may need to be customized.
The order of operations is controlled largely by the pipeline function full_import which is the typical use case method for importing data. That function will check that the import file(s) include requirements and recommendations as defined in /config/NIST_import_requirements.json which is a JSON list of expected elements and headers within each element and whether the elements are required. When using the NTA MRT format and process to import data the default arguments to this function and the import map should not be changed, but flexibility is supported by full_import having a nearly exhaustive list of parameters passed to underlying functions to resolve each database node in the required order (contributors, methods, descriptions, samples, chromatography, quality control, peaks, compounds, and fragments; see SQL Nodes in Technical Details for more details about schema nodes); parameters are passed largely by name matches for underlying functions using do.call. The import process is only available from the console, provides logging (if enabled) throughout, and fully supports batch imports from a list of import files read in via jsonlite::fromJSON(readr::read_file(X)) where X is a vector of file paths. Files may alternatively be imported one at a time directly from JSON files using the file_name parameter and leaving the import_object parameter as NULL. A live connection to the database is required, and when additional information is needed (e.g. to resolve or add unknown controlled table entries), users will be prompted at the command line during the process.
Alternatively, data can be imported when a database is built or rebuilt from comma-separated value (CSV) files. This process is not likely amenable to many projects as it requires data indices be prepopulated and accurately cross-linked across CSV files, with one CVS file for each database table being populated; this should be considered if data are already in a database-like format and can be easily cross-linked, in which case only the table and column mappings need be solved. Several such files are used to populate a “clean” database install with certain controlled vocabulary and reference tables (see files /config/populate_common.sql and the /config/data/ directory). Contact these authors for assistance with using the NTA MRT and msconvert process, or conversion of data into the DIMSpec schema if you feel a project’s data would be amenable to the database structure described in this document.
Ending Your Session
Unclosed database connections can have unintended consequences. Generally, connections to the database during a session should be managed with manage_connection which allows for both disconnect and reconnect (to flush the WAL and establish a new connection). The API server will need to be spun down separately using stop_api. Alternatively, and to preserve any data frame objects that may have been created as external pointers (i.e. as dplyr tbls), when users finish with their connection needs they may use the convenience function close_up_shop. Connections may not flush completely in all cases. If users notice the -shm and -wal files are still open in the directory, the best way to flush them is to establish a new connection and then disconnect from it, using either manage_connection or DBI::dbConnect/DBI::dbDisconnect.
Updating the Schema
At the time this book was written, the schema should be well defined for most use cases. Extensions can however be added at any time to suit project-specific needs. To avoid data loss, it is recommended that any table extensions be performed directly in SQL and those commands saved to an SQL script. Views can be added freely as required. If users of this database framework apply any schema extensions, the authors would be interested in learning about both the need and the implementation so it may be evaluated for inclusion in future versions.
This concludes the User Guide for the Database Infrastructure for Mass Spectrometry. The following section contains technical details about the implementation and user customization.
References
Any mention of commercial products within NIST web pages is for information only; it does not imply recommendation or endorsement by NIST.↩︎
This release was tested on a fresh VMWare build of Ubuntu 20.04 LTS which carries several additional system requirements. Prior to running DIMSpec, install or make sure the following are available using:
apt install -y build-essential libcurl4-openssl-dev libxml2-dev zlib1g-dev libssl-dev libsodium-dev ffmpeg libtiff-dev libpng-dev libblas-dev liblapack-dev libarpack2-dev gfortran libcairo2-dev libx11-dev libharfbuzz-dev libfribidi-dev libudunits2-dev libgeos-dev libgdal-dev libfftw3-3 libmagick++-devAfter following the R installation instructions for Ubuntu, ensure additional requirements using:apt install -y –no-install-recommends r-cran-tidyverse r-cran-shiny↩︎