examples.cahnHilliard.mesh2D¶
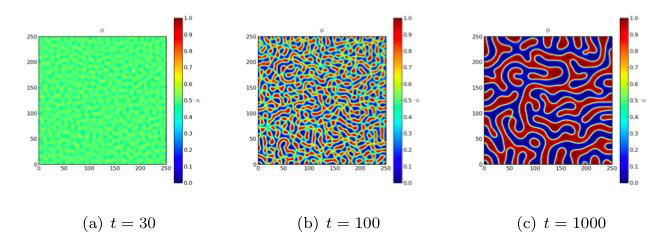
The spinodal decomposition phenomenon is a spontaneous separation of an initially homogeneous mixture into two distinct regions of different properties (spin-up/spin-down, component A/component B). It is a “barrierless” phase separation process, such that under the right thermodynamic conditions, any fluctuation, no matter how small, will tend to grow. This is in contrast to nucleation, where a fluctuation must exceed some critical magnitude before it will survive and grow. Spinodal decomposition can be described by the “Cahn-Hilliard” equation (also known as “conserved Ginsberg-Landau” or “model B” of Hohenberg & Halperin)
where \(\phi\) is a conserved order parameter, possibly representing alloy composition or spin. The double-well free energy function \(f = (a^2/2) \phi^2 (1 - \phi)^2\) penalizes states with intermediate values of \(\phi\) between 0 and 1. The gradient energy term \(\epsilon^2 \nabla^2\phi\), on the other hand, penalizes sharp changes of \(\phi\). These two competing effects result in the segregation of \(\phi\) into domains of 0 and 1, separated by abrupt, but smooth, transitions. The parameters \(a\) and \(\epsilon\) determine the relative weighting of the two effects and \(D\) is a rate constant.
We can simulate this process in FiPy with a simple script:
>>> from fipy import CellVariable, Grid2D, GaussianNoiseVariable, TransientTerm, DiffusionTerm, ImplicitSourceTerm, LinearLUSolver, Viewer, DefaultSolver
>>> from fipy.tools import numerix
(Note that all of the functionality of NumPy is imported along with FiPy, although much is augmented for FiPy's needs.)
>>> if __name__ == "__main__":
... nx = ny = 1000
... else:
... nx = ny = 20
>>> mesh = Grid2D(nx=nx, ny=ny, dx=0.25, dy=0.25)
>>> phi = CellVariable(name=r"$\phi$", mesh=mesh)
We start the problem with random fluctuations about \(\phi = 1/2\)
>>> phi.setValue(GaussianNoiseVariable(mesh=mesh,
... mean=0.5,
... variance=0.01))
FiPy doesn’t plot or output anything unless you tell it to:
>>> if __name__ == "__main__":
... viewer = Viewer(vars=(phi,), datamin=0., datamax=1.)
For FiPy, we need to perform the partial derivative
\(\partial f/\partial \phi\)
manually and then put the equation in the canonical
form by decomposing the spatial derivatives
so that each Term is of a single, even order:
FiPy would automatically interpolate
D * a**2 * (1 - 6 * phi * (1 - phi))
onto the faces, where the diffusive flux is calculated, but we obtain
somewhat more accurate results by performing a linear interpolation from
phi at cell centers to PHI at face centers.
Some problems benefit from non-linear interpolations, such as harmonic or
geometric means, and FiPy makes it easy to obtain these, too.
>>> PHI = phi.arithmeticFaceValue
>>> D = a = epsilon = 1.
>>> eq = (TransientTerm()
... == DiffusionTerm(coeff=D * a**2 * (1 - 6 * PHI * (1 - PHI)))
... - DiffusionTerm(coeff=(D, epsilon**2)))
>>> import fipy.solvers.solver
>>> if fipy.solvers.solver_suite in ['petsc']:
... solver = DefaultSolver(precon=None)
... elif fipy.solvers.solver_suite in ['trilinos']:
... solver = LinearLUSolver()
... else:
... solver = DefaultSolver()
Because the evolution of a spinodal microstructure slows with time, we use exponentially increasing time steps to keep the simulation “interesting”. The FiPy user always has direct control over the evolution of their problem.
>>> dexp = -5
>>> elapsed = 0.
>>> duration = 1000.
>>> while elapsed < duration:
... dt = min(100, numerix.exp(dexp))
... elapsed += dt
... dexp += 0.01
... eq.solve(phi, dt=dt, solver=solver)
... if __name__ == "__main__":
... viewer.plot()
... elif (max(phi.globalValue) > 0.7) and (min(phi.globalValue) < 0.3) and elapsed > 10.:
... break
>>> print((max(phi.globalValue) > 0.7) and (min(phi.globalValue) < 0.3))
True
 FiPy
FiPy