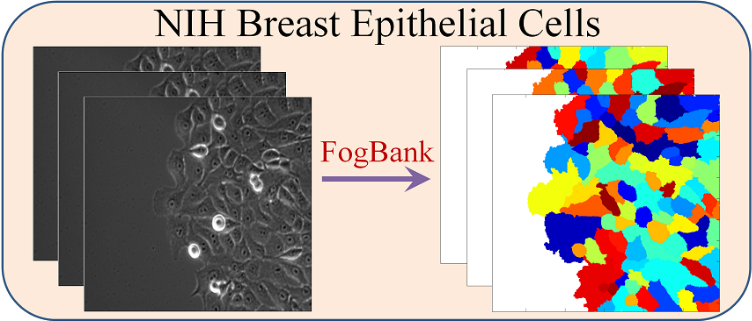
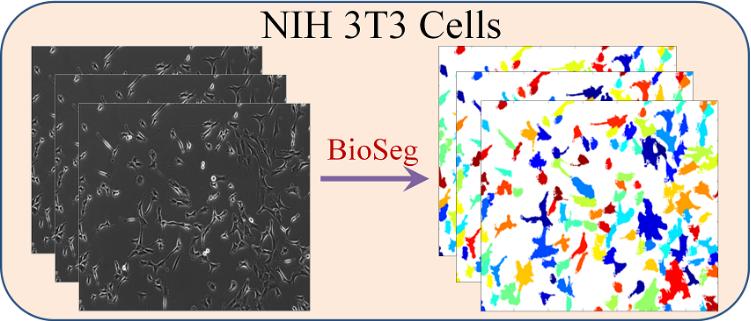
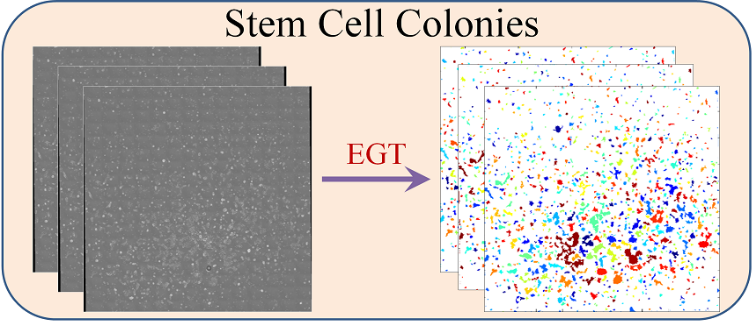
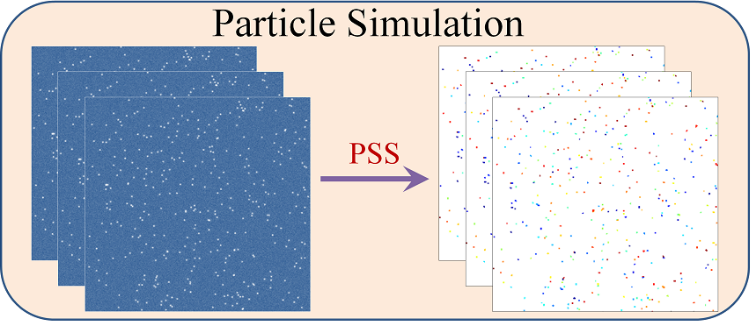
What is Lineage-Mapper?
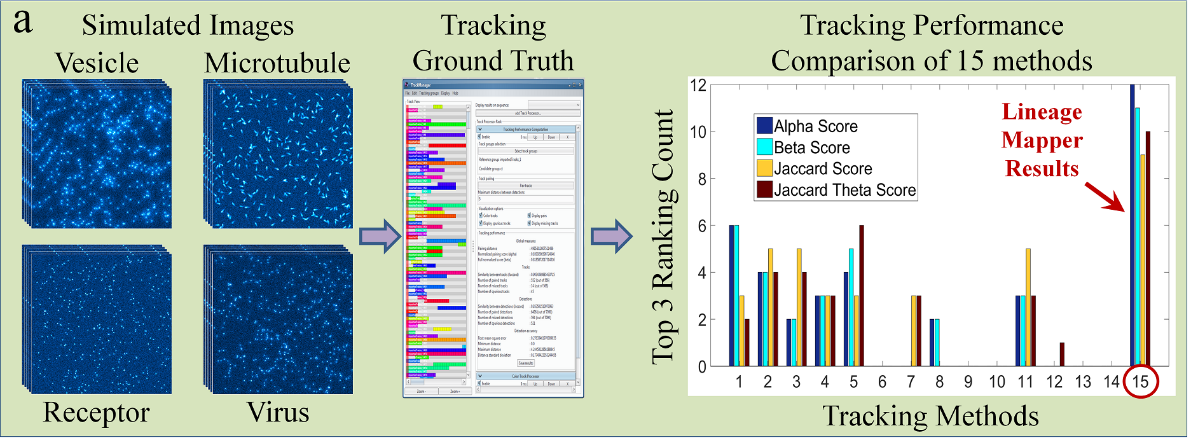
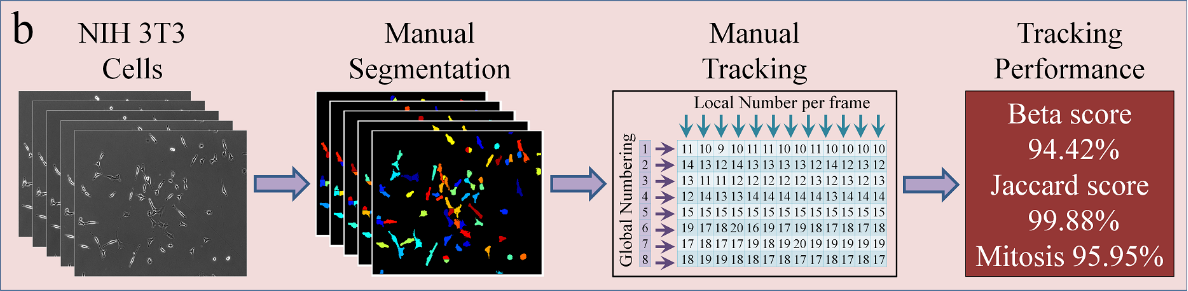
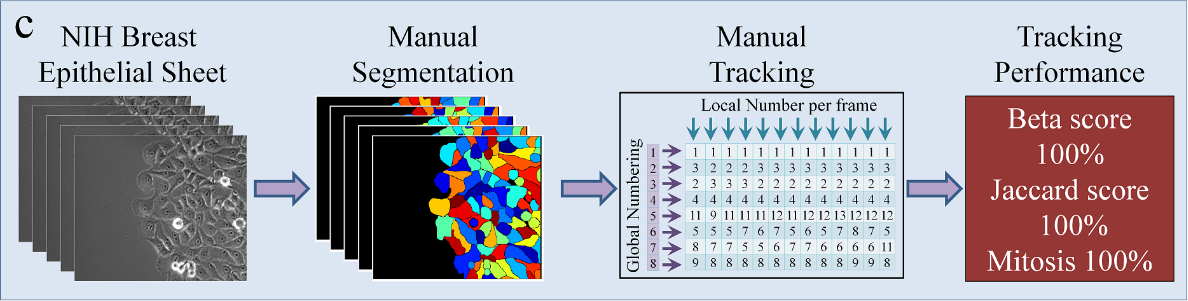
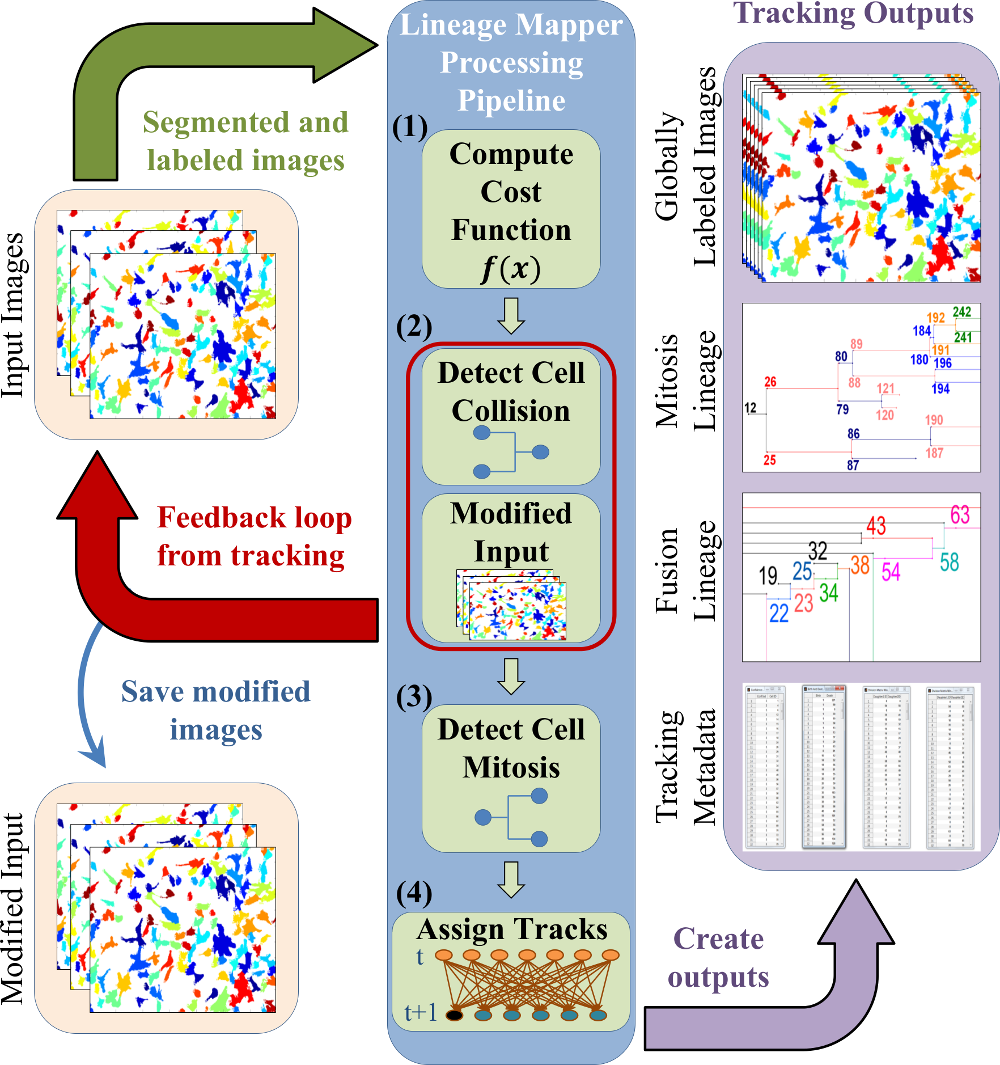
Lineage Mapper is an open-source, highly accurate, overlap-based cell tracking system for time-lapse images of biological cells, colonies, and particles. Lineage Mapper tracks objects independently of the segmentation method, detects mitosis in confluence, separates cell clumps mistakenly segmented as a single cell, provides accuracy and scalability even on terabyte-sized datasets, and creates division and/or fusion lineages. Lineage Mapper has been tested and validated on multiple biological and simulated problems.
Staff:
Publications:
J. Chalfoun, M. Majurski, A. Dima, M. Halter, K. Bhadriraju, and M. Brady, “Lineage mapper: A versatile cell and particle tracker,” Scientific Reports, vol. 6, October, 2016, DOI: 10.1038/srep36984
(view article)
J. Chalfoun, A. Cardone, and A. Dima, “Overlap-based cell tracker,” J. Research Natl. Inst. Stand. Technol., vol. 115, no. 6, p. 477, Nov. 2010
(download pdf)
(view article)